Analysis of mupirocin resistance and fitness in Staphylococcus aureus by molecular genetic and structural modeling techniques
- PMID: 15504866
- PMCID: PMC525403
- DOI: 10.1128/AAC.48.11.4366-4376.2004
Analysis of mupirocin resistance and fitness in Staphylococcus aureus by molecular genetic and structural modeling techniques
Abstract
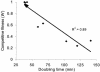
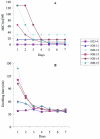
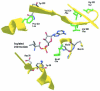
Chromosomal resistance to mupirocin in clinical isolates of Staphylococcus aureus arises from V(588)F or V(631)F mutations in isoleucyl-tRNA synthetase (IRS). Whether these are the only IRS mutations that confer mupirocin resistance or simply those that survive in the clinic is unknown. Mupirocin-resistant mutants of S. aureus 8325-4 were therefore generated to examine their ileS genotypes and the in vitro and in vivo fitness costs associated with them before and after compensatory evolution. Most spontaneous first-step mupirocin-resistant mutants carried V(588)F or V(631)F mutations in IRS, but a new mutation (G(593)V) was also identified. Second-step mutants carried combinations of previously identified IRS mutations (e.g., V(588)F/V(631)F and G(593)V/V(631)F), but additional combinations also occurred involving novel mutations (R(816)C, H(67)Q, and F(563)L). First-step mupirocin-resistant mutants were not associated with substantial fitness costs, a finding that is consistent with the occurrence of V(588)F or V(631)F mutations in the IRS of clinical strains. Second-step mutants were unfit, but fitness could be restored by subculture in the absence of mupirocin. In most cases, this was the result of compensatory mutations that also suppressed mupirocin resistance (e.g., A(196)V, E(190)K, and E(195)K), despite retention of the original mutations conferring resistance. Structural explanations for mupirocin resistance and loss of fitness were obtained by molecular modeling of mutated IRS enzymes, which provided data on mupirocin binding and interaction with the isoleucyl-AMP reactive intermediate.
Figures

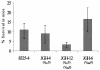




References
-
- Andersson, D. I., and B. R. Levin. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489-493. - PubMed
-
- Brown, M. J. B., L. M. Mensah, M. L. Doyle, N. J. P. Broom, N. Osbourne, A. K. Forrest, C. M. Richardson, P. J. O'Hanlon, and A. J. Pope. 2000. Rational design of femtomolar inhibitors of isoleucyl tRNA synthetase from a binding model for pseudomonic acid-A. Biochemistry 39:6003-6011. - PubMed
-
- Cookson, B. D. 1998. The emergence of mupirocin resistance: a challenge to infection control and antibiotic prescribing practice. J. Antimicrob. Chemother. 41:11-18. - PubMed
-
- Decousser, J. W., P. Pina, J. C. Ghnassia, J. P. Bedos, and P. Y. Allouch. 2003. First report of clinical and microbiological failure in the eradication of glycopeptide-intermediate methicillin-resistant Staphylococcus aureus carriage by mupirocin. Eur. J. Clin. Microbiol. 22:318-319. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

