Severe acute respiratory syndrome coronavirus 3C-like proteinase N terminus is indispensable for proteolytic activity but not for enzyme dimerization. Biochemical and thermodynamic investigation in conjunction with molecular dynamics simulations
- PMID: 15507456
- PMCID: PMC7982548
- DOI: 10.1074/jbc.M408211200
Severe acute respiratory syndrome coronavirus 3C-like proteinase N terminus is indispensable for proteolytic activity but not for enzyme dimerization. Biochemical and thermodynamic investigation in conjunction with molecular dynamics simulations
Abstract
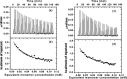
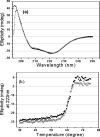
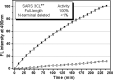
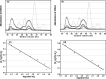
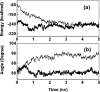
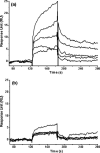
Severe acute respiratory syndrome (SARS) coronavirus is a novel human coronavirus and is responsible for SARS infection. SARS coronavirus 3C-like proteinase (SARS 3CL(pro)) plays key roles in viral replication and transcription and is an attractive target for anti-SARS drug discovery. In this report, we quantitatively characterized the dimerization features of the full-length and N-terminal residues 1-7 deleted SARS 3CL(pro)s by using glutaraldehyde cross-linking SDS-PAGE, size-exclusion chromatography, and isothermal titration calorimeter techniques. Glutaraldehyde cross-linking SDS-PAGE and size-exclusion chromatography results show that, similar to the full-length SARS 3CL(pro), the N-terminal deleted SARS 3CL(pro) still remains a dimer/monomer mixture within a wide range of protein concentrations. Isothermal titration calorimeter determinations indicate that the equilibrium dissociation constant (K(d)) of the N-terminal deleted proteinase dimer (262 microm) is very similar to that of the full-length proteinase dimer (227 microm). Enzymatic activity assay using the fluorescence resonance energy transfer method reveals that N-terminal deletion results in almost complete loss of enzymatic activity for SARS 3CL(pro). Molecular dynamics and docking simulations demonstrate the N-terminal deleted proteinase dimer adopts a state different from that of the full-length proteinase dimer, which increases the angle between the two protomers and reduces the binding pocket that is not beneficial to the substrate binding. This conclusion is verified by the surface plasmon resonance biosensor determination, indicating that the model substrate cannot bind to the N-terminal deleted proteinase. These results suggest the N terminus is not indispensable for the proteinase dimerization but may fix the dimer at the active state and is therefore vital to enzymatic activity.
Figures

References
-
- Kathryn V.H. J. Clin. Investig. 2002;111:1605–1609.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

