Human fatty acid synthase: structure and substrate selectivity of the thioesterase domain
- PMID: 15507492
- PMCID: PMC524853
- DOI: 10.1073/pnas.0406901101
Human fatty acid synthase: structure and substrate selectivity of the thioesterase domain
Abstract
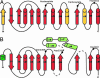
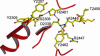
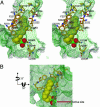
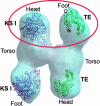
Human fatty acid synthase is a large homodimeric multifunctional enzyme that synthesizes palmitic acid. The unique carboxyl terminal thioesterase domain of fatty acid synthase hydrolyzes the growing fatty acid chain and plays a critical role in regulating the chain length of fatty acid released. Also, the up-regulation of human fatty acid synthase in a variety of cancer makes the thioesterase a candidate target for therapeutic treatment. The 2.6-A resolution structure of human fatty acid synthase thioesterase domain reported here is comprised of two dissimilar subdomains, A and B. The smaller subdomain B is composed entirely of alpha-helices arranged in an atypical fold, whereas the A subdomain is a variation of the alpha/beta hydrolase fold. The structure revealed the presence of a hydrophobic groove with a distal pocket at the interface of the two subdomains, which constitutes the candidate substrate binding site. The length and largely hydrophobic nature of the groove and pocket are consistent with the high selectivity of the thioesterase for palmitoyl acyl substrate. The structure also set the identity of the Asp residue of the catalytic triad of Ser, His, and Asp located in subdomain A at the proximal end of the groove.
Figures


References
-
- Wakil, S. J. (1989) Biochemistry 28, 4523-4530. - PubMed
-
- Smith, S. (1994) FASEB J. 8, 1248-1259. - PubMed
-
- Must, A., Spadano, J., Coakley, E. H., Field, A. E., Colditz, G. & Dietz, W. H. (1999) J. Am. Med. Assoc. 282, 1523-1529. - PubMed
-
- Alo, P. L., Visca, P., Marci, A., Mangoni, A., Botti, C. & Di Tondo, U. (1996) Cancer 77, 474-482. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

