A retroviral promoter and a cellular enhancer define a bipartite element which controls env ERVWE1 placental expression
- PMID: 15507602
- PMCID: PMC525085
- DOI: 10.1128/JVI.78.22.12157-12168.2004
A retroviral promoter and a cellular enhancer define a bipartite element which controls env ERVWE1 placental expression
Abstract
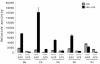
The HERV-W family contains hundreds of loci diversely expressed in several physiological and pathological contexts. A unique locus termed ERVWE1 encodes an envelope glycoprotein (syncytin) involved in hominoid placental physiology. Here we show that syncytin expression is regulated by a bipartite element consisting of a cyclic AMP (cAMP)-inducible long terminal repeat (LTR) retroviral promoter adjacent to a cellular enhancer conferring a high level of expression and placental tropism. Deletion mutant analysis showed that the ERVWE1 5' LTR contains binding sites essential for basal placental activity in the region from positions +1 to +125. The region from positions +125 to +310 represents a cAMP-responsive core HERV-W promoter active in all cell types. Site-directed mutagenesis analysis highlighted the complexity of U3 regulation. ERVWE1 placenta-specific positive (e.g., T240) and negative (e.g., G71) regulatory sites were identified, as were essential sites required for basic activity (e.g., A247). The flanking sequences of the ERVWE1 provirus contain several putative regulatory elements. The upstream HERV-H and HERV-P LTRs were found to be inactive. Conversely, the 436-bp region located between the HERV-P LTR and ERVWE1 was shown to be an upstream regulatory element (URE) which is significantly active in placenta cells. This URE acts as a tissue-specific enhancer. Genetic and functional analyses of hominoid UREs revealed large differences between UREs of members of the Hominidae and the Hylobatidae. These data allowed the identification of a positive regulatory region from positions -436 to -128, a mammalian apparent LTR retrotransposon negative regulatory region from positions -128 to -67, and a trophoblast-specific enhancer (TSE) from positions -67 to -35. Putative AP-2, Sp-1, and GCMa binding sites are essential constituents of the 33-bp TSE.
Figures

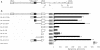




Similar articles
-
Natural history of the ERVWE1 endogenous retroviral locus.Retrovirology. 2005 Sep 22;2:57. doi: 10.1186/1742-4690-2-57. Retrovirology. 2005. PMID: 16176588 Free PMC article.
-
The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology.Proc Natl Acad Sci U S A. 2004 Feb 10;101(6):1731-6. doi: 10.1073/pnas.0305763101. Epub 2004 Feb 2. Proc Natl Acad Sci U S A. 2004. PMID: 14757826 Free PMC article.
-
Evidence of selection on the domesticated ERVWE1 env retroviral element involved in placentation.Mol Biol Evol. 2004 Oct;21(10):1895-901. doi: 10.1093/molbev/msh206. Epub 2004 Jul 14. Mol Biol Evol. 2004. PMID: 15254254
-
Expression and regulation of human endogenous retrovirus W elements.APMIS. 2016 Jan-Feb;124(1-2):52-66. doi: 10.1111/apm.12478. APMIS. 2016. PMID: 26818262 Review.
-
[Retroviral inheritance in man].J Soc Biol. 2004;198(4):399-412. J Soc Biol. 2004. PMID: 15969347 Review. French.
Cited by
-
Long-Term Endurance Exercise in Humans Stimulates Cell Fusion of Myoblasts along with Fusogenic Endogenous Retroviral Genes In Vivo.PLoS One. 2015 Jul 8;10(7):e0132099. doi: 10.1371/journal.pone.0132099. eCollection 2015. PLoS One. 2015. PMID: 26154387 Free PMC article.
-
Transposable element-derived sequences in vertebrate development.Mob DNA. 2021 Jan 6;12(1):1. doi: 10.1186/s13100-020-00229-5. Mob DNA. 2021. PMID: 33407840 Free PMC article. Review.
-
Reduced syncytin-1 expression levels in placental syndromes correlates with epigenetic hypermethylation of the ERVW-1 promoter region.PLoS One. 2013;8(2):e56145. doi: 10.1371/journal.pone.0056145. Epub 2013 Feb 14. PLoS One. 2013. PMID: 23457515 Free PMC article.
-
Impaired cell fusion and differentiation in placentae from patients with intrauterine growth restriction correlate with reduced levels of HERV envelope genes.J Mol Med (Berl). 2010 Nov;88(11):1143-56. doi: 10.1007/s00109-010-0656-8. Epub 2010 Jul 28. J Mol Med (Berl). 2010. PMID: 20664994
-
HERV Envelope Proteins: Physiological Role and Pathogenic Potential in Cancer and Autoimmunity.Front Microbiol. 2018 Mar 14;9:462. doi: 10.3389/fmicb.2018.00462. eCollection 2018. Front Microbiol. 2018. PMID: 29593697 Free PMC article. Review.
References
-
- Anson-Cartwright, L., K. Dawson, D. Holmyard, S. J. Fisher, R. A. Lazzarini, and J. C. Cross. 2000. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat. Genet. 25:311-314. - PubMed
-
- Ben Zimra, M., M. Koler, and J. Orly. 2002. Transcription of cholesterol side-chain cleavage cytochrome P450 in the placenta: activating protein-2 assumes the role of steroidogenic factor-1 by binding to an overlapping promoter element. Mol. Endocrinol. 16:1864-1880. - PubMed
-
- Bi, S., O. Gavrilova, D. W. Gong, M. M. Mason, and M. Reitman. 1997. Identification of a placental enhancer for the human leptin gene. J. Biol. Chem. 272:30583-30588. - PubMed
-
- Bieche, I., A. Laurent, I. Laurendeau, L. Duret, Y. Giovangrandi, J. L. Frendo, M. Olivi, J. L. Fausser, D. Evain-Brion, and M. Vidaud. 2003. Placenta-specific INSL4 expression is mediated by a human endogenous retrovirus element. Biol. Reprod. 68:1422-1429. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

