Identification and characterization of persistent human erythrovirus infection in blood donor samples
- PMID: 15507603
- PMCID: PMC525065
- DOI: 10.1128/JVI.78.22.12169-12178.2004
Identification and characterization of persistent human erythrovirus infection in blood donor samples
Abstract
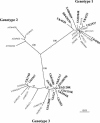
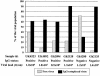
The presence of human erythrovirus DNA in 2,440 blood donations from the United Kingdom and sub-Saharan Africa (Ghana, Malawi, and South Africa) was screened. Sensitive qualitative and real-time quantitative PCR assays revealed a higher prevalence of persistent infection with the simultaneous presence of immunoglobulin G (IgG) and viral DNA (0.55 to 1.3%) than previously reported. This condition was characterized by a low viral load (median, 558 IU/ml; range, 42 to 135,000 IU/ml), antibody-complexed virus, free specific IgG, and potentially infectious free virus. Human erythrovirus genotype 1 (formerly parvovirus B19) was prevalent in the United Kingdom, Malawi, and South Africa. In contrast, only human erythrovirus genotype 3 (erythrovirus variant V9) was prevalent in Ghana. Genotype 3 had considerable genetic diversity, clustering in two probable subtypes. Genotype 1-based antibody assays failed to detect 38.5% of Ghanaian samples containing antibodies to genotype 3 virus but did not fail to detect cases of persistent infection. This study indicates a potential African origin of genotype 3 human erythrovirus and considerable shortcomings in the tools currently used to diagnose erythrovirus infection.
Figures


Comment in
-
Evidence of serological cross-reactivity between genotype 1 and genotype 3 erythrovirus infections.J Virol. 2005 Apr;79(8):5238-9; author reply 5239. doi: 10.1128/JVI.79.8.5238-5239.2005. J Virol. 2005. PMID: 15795309 Free PMC article. No abstract available.
References
-
- Allain, J.-P., D. Candotti, K. Soldan, F. Sarkodie, B. Phelps, C. Giachetti, V. Shyamala, F. Yeboah, M. Anokwa, S. Owusu-Ofori, and O. Opare-Sem. 2003. The risk of hepatitis B virus infection by transfusion in Kumasi, Ghana. Blood 101:2419-2425. - PubMed
-
- Aubin, J.-T., C. Defer, M. Vidaud, M. Maniez Montreuil, and B. Flan. 2000. Large-scale screening for human parvovirus B19 DNA by PCR: application to the quality control of plasma for fractionation. Vox Sang. 78:7-12. - PubMed
-
- Blumel, J., I. Schmidt, W. Effenberger, H. Seitz, H. Willkommen, H. H. Brackmann, J. Lower, and A. M. Eis-Hubinger. 2002. Parvovirus B19 transmission by heat-treated clotting factor concentrates. Transfusion 42:1473-1481. - PubMed
-
- Brechot, C., V. Thiers, D. Kremsdorf, B. Nalpas, S. Pol, and P. Paterlini-Brechot. 2001. Persistent hepatitis B virus infection in subjects without hepatitis B surface antigen: clinically significant or purely “occult”? Hepatology 34:194-203. - PubMed
-
- Brown, K. E., N. S. Young, B. M. Alving, and L. H. Barbosa. 2001. Parvovirus B19: implications for transfusion medicine. Summary of a workshop. Transfusion 41:130-135. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

