The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor
- PMID: 15507608
- PMCID: PMC525082
- DOI: 10.1128/JVI.78.22.12218-12224.2004
The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor
Abstract
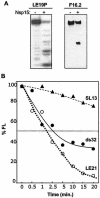
Nonstructural protein 15 (Nsp15) of the severe acute respiratory syndrome coronavirus (SARS-CoV) produced in Escherichia coli has endoribonuclease activity that preferentially cleaved 5' of uridylates of RNAs. Blocking either the 5' or 3' terminus did not affect cleavage. Double- and single-stranded RNAs were both substrates for Nsp15 but with different kinetics for cleavage. Mn(2+) at 2 to 10 mM was needed for optimal endoribonuclease activity, but Mg(2+) and several other divalent metals were capable of supporting only a low level of activity. Concentrations of Mn(2+) needed for endoribonuclease activity induced significant conformation change(s) in the protein, as measured by changes in tryptophan fluorescence. A similar endoribonucleolytic activity was detected for the orthologous protein from another coronavirus, demonstrating that the endoribonuclease activity of Nsp15 may be common to coronaviruses. This work presents an initial biochemical characterization of a novel coronavirus endoribonuclease.
Figures


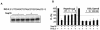


References
-
- Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. - PubMed
-
- Caffarelli, E., L. Maggi, A. Fatica, J. Jiricny, and I. Bozzoni. 1997. A novel Mn2+-dependent ribonuclease that functions in U16 SnoRNA processing in X. laevis. Biochem. Biophys. Res. Commun. 233:514-517. - PubMed
-
- Enserink, M. 2003. Clues to the animal origins of SARS. Science 300:1351. - PubMed
-
- Goedken, E. R., and S. Marqusee. 2001. Co-crystal of Escherichia coli RNase HI with Mn2+ ions reveals two divalent metals bound in the active site. J. Biol. Chem. 276:7266-7271. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

