Murine T cells potently restrict human immunodeficiency virus infection
- PMID: 15507641
- PMCID: PMC525105
- DOI: 10.1128/JVI.78.22.12537-12547.2004
Murine T cells potently restrict human immunodeficiency virus infection
Abstract
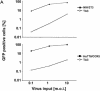
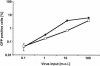
Development of a mouse model for human immunodeficiency virus type 1 (HIV-1) infection has advanced through the progressive identification of host cell factors required for HIV-1 replication. Murine cells lack HIV-1 receptor molecules, do not support efficient viral gene expression, and lack factors necessary for the assembly and release of virions. Many of these blocks have been described using mouse fibroblast cell lines. Here we identify a postentry block to HIV-1 infection in mouse T-cell lines that has not been detected in mouse fibroblasts. While murine fibroblastic lines are comparable to human T-cell lines in permissivity to HIV-1 transduction, infection of murine T cells is 100-fold less efficient. Virus entry occurs efficiently in murine T cells. However, reduced efficiency of the completion of reverse transcription and nuclear transfer of the viral preintegration complex are observed. Although this block has similarities to the restriction of murine retroviruses by Fv1, there is no correlation of HIV-1 susceptibility with cellular Fv1 genotypes. In addition, the block to HIV-1 infection in murine T-cell lines cannot be saturated by a high virus dose. Further studies of this newly identified block may lend insight into the early events of retroviral replication and reveal new targets for antiretroviral interventions.
Figures







References
-
- Aagaard, L., J. G. Mikkelsen, S. Warming, M. Duch, and F. S. Pedersen. 2002. Fv1-like restriction of N-tropic replication-competent murine leukaemia viruses in mCAT-1-expressing human cells. J. Gen. Virol. 83:439-442. - PubMed
-
- Aaronson, S. A., and G. J. Todaro. 1968. Development of 3T3-like lines from Balb-c mouse embryo cultures: transformation susceptibility to SV40. J. Cell Physiol. 72:141-148. - PubMed
-
- Benjers, B. M., R. H. Bassin, A. Rein, B. I. Gerwin, and G. Duran-Troise. 1979. Mechanism of restriction of murine leukemia viruses varies between different strains of Fv-1n mice. Int. J. Cancer 24:600-607. - PubMed
-
- Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

