The nucleoprotein is required for efficient coronavirus genome replication
- PMID: 15507657
- PMCID: PMC525053
- DOI: 10.1128/JVI.78.22.12683-12688.2004
The nucleoprotein is required for efficient coronavirus genome replication
Abstract
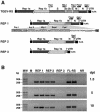
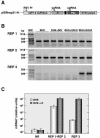
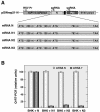
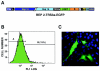
The construction of a set of transmissible gastroenteritis coronavirus (TGEV)-derived replicons as bacterial artificial chromosomes is reported. These replicons were generated by sequential deletion of nonessential genes for virus replication, using a modified TGEV full-length cDNA clone containing unique restriction sites between each pair of consecutive genes. Efficient activity of TGEV replicons was associated with the presence of the nucleoprotein provided either in cis or in trans. TGEV replicons were functional in several cell lines, including the human cell line 293T, in which no or very low cytopathic effect was observed, and expressed high amounts of heterologous protein.
Figures
References
-
- Delmas, B., J. Gelfi, H. Sjöström, O. Noren, and H. Laude. 1994. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv. Exp. Med. Biol. 342:293-298. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

