Measurement of relative copy number of CDKN2A/ARF and CDKN2B in bladder cancer by real-time quantitative PCR and multiplex ligation-dependent probe amplification
- PMID: 15507675
- PMCID: PMC1867481
- DOI: 10.1016/S1525-1578(10)60532-6
Measurement of relative copy number of CDKN2A/ARF and CDKN2B in bladder cancer by real-time quantitative PCR and multiplex ligation-dependent probe amplification
Abstract
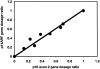
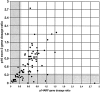
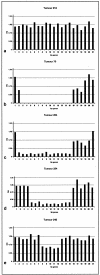
Many tumors have large homozygous deletions of the CDKN2A locus (encoding p14(ARF) and p16) and of CDKN2B (p15). Our aim was to determine which gene is the major target in bladder cancer. We used quantitative real-time PCR (RTQ-PCR) to determine copy number of p15, of p14(ARF) exon 1beta, and p16 exon 2 in 22 tumor cell lines and 83 bladder tumors, some of which had been assessed previously by duplex PCR. Titration experiments showed that homozygous deletion could be detected in the presence of up to 30% normal DNA. Results for cell lines were compatible with previous cytogenetic analyses. Ten cell lines and 32 tumors (38.5%) had homozygous deletion of at least one target. Thirteen tumors (15.7%) had deletion of all three targets. Two tumors had deletion of p14(ARF) exon 1beta alone and four of p16 exon 2 alone. RTQ-PCR detected more homozygous deletions than duplex PCR. Finally we used a multiplex ligation-dependent probe amplification kit to provide independent confirmation of results. We conclude that with appropriate controls RTQ-PCR is a sensitive and robust method to detect copy number changes in tumors even in the presence of contaminating normal cell DNA.
Figures



Similar articles
-
Fine-mapping loss of gene architecture at the CDKN2B (p15INK4b), CDKN2A (p14ARF, p16INK4a), and MTAP genes in head and neck squamous cell carcinoma.Arch Otolaryngol Head Neck Surg. 2006 Apr;132(4):409-15. doi: 10.1001/archotol.132.4.409. Arch Otolaryngol Head Neck Surg. 2006. PMID: 16618910
-
Detecting homozygous deletions in the CDKN2A(p16(INK4a))/ARF(p14(ARF)) gene in urinary bladder cancer using real-time quantitative PCR.Clin Cancer Res. 2003 Jan;9(1):235-42. Clin Cancer Res. 2003. PMID: 12538475
-
Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas.Am J Pathol. 2001 Aug;159(2):661-9. doi: 10.1016/S0002-9440(10)61737-3. Am J Pathol. 2001. PMID: 11485924 Free PMC article.
-
Molecular analysis of P16(Ink4)/CDKN2 and P15(INK4B)/MTS2 genes in primary human testicular germ cell tumors.J Urol. 1998 May;159(5):1725-30. doi: 10.1097/00005392-199805000-00101. J Urol. 1998. PMID: 9554401
-
Homozygous deletions at chromosome 9p21 and mutation analysis of p16 and p15 in microdissected primary non-small cell lung cancers.Clin Cancer Res. 1995 Jul;1(7):687-90. Clin Cancer Res. 1995. PMID: 9816033
Cited by
-
CDKN2A as transcriptomic marker for muscle-invasive bladder cancer risk stratification and therapy decision-making.Sci Rep. 2018 Sep 26;8(1):14383. doi: 10.1038/s41598-018-32569-x. Sci Rep. 2018. PMID: 30258198 Free PMC article.
-
Chromosome 9p21 gene copy number and prognostic significance of p16 in ESFT.Br J Cancer. 2007 Jun 18;96(12):1914-23. doi: 10.1038/sj.bjc.6603819. Epub 2007 May 29. Br J Cancer. 2007. PMID: 17533400 Free PMC article.
-
Alterations of RB1 gene in embryonal and alveolar rhabdomyosarcoma: special reference to utility of pRB immunoreactivity in differential diagnosis of rhabdomyosarcoma subtype.J Cancer Res Clin Oncol. 2008 Oct;134(10):1097-103. doi: 10.1007/s00432-008-0385-3. Epub 2008 Apr 2. J Cancer Res Clin Oncol. 2008. PMID: 18386058 Free PMC article.
-
Urothelial tumor initiation requires deregulation of multiple signaling pathways: implications in target-based therapies.Carcinogenesis. 2012 Apr;33(4):770-80. doi: 10.1093/carcin/bgs025. Epub 2012 Jan 27. Carcinogenesis. 2012. PMID: 22287562 Free PMC article.
-
Mapping of the chromosomal amplification 1p21-22 in bladder cancer.BMC Res Notes. 2014 Aug 18;7:547. doi: 10.1186/1756-0500-7-547. BMC Res Notes. 2014. PMID: 25135188 Free PMC article.
References
-
- Tsai YC, Nichols PW, Hiti AL, Williams Z, Skinner DG, Jones PA. Allelic losses of chromosomes 9, 11, and 17 in human bladder cancer. Cancer Res. 1990;50:44–47. - PubMed
-
- Cairns P, Shaw ME, Knowles MA. Initiation of bladder cancer may involve deletion of a tumour-suppressor gene on chromosome 9. Oncogene. 1993;8:1083–1085. - PubMed
-
- Cairns P, Shaw ME, Knowles MA. Preliminary mapping of the deleted region of chromosome 9 in bladder cancer. Cancer Res. 1993;53:1230–1232. - PubMed
-
- Miyao N, Tsai YC, Lerner SP, Olumi AF, Spruck CH, III, Gonzalez-Zulueta M, Nichols PW, Skinner DG, Jones PA. Role of chromosome 9 in human bladder cancer. Cancer Res. 1993;53:4066–4070. - PubMed
-
- Habuchi T, Ogawa O, Kakehi Y, Ogura K, Koshiba M, Hamazaki S, Takahashi R, Sugiyama T, Yoshida O. Accumulated allelic losses in the development of invasive urothelial cancer. Int J Cancer. 1993;53:579–584. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

