In silico evidence for DNA polymerase-beta's substrate-induced conformational change
- PMID: 15507687
- PMCID: PMC1304780
- DOI: 10.1529/biophysj.104.040915
In silico evidence for DNA polymerase-beta's substrate-induced conformational change
Abstract
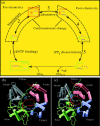
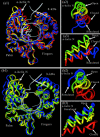
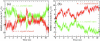
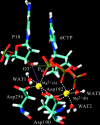
Structural information for mammalian DNA pol-beta combined with molecular and essential dynamics studies have provided atomistically detailed views of functionally important conformational rearrangements that occur during DNA repair and replication. This conformational closing before the chemical reaction is explored in this work as a function of the bound substrate. Anchors for our study are available in crystallographic structures of the DNA pol-beta in "open" (polymerase bound to gapped DNA) and "closed" (polymerase bound to gapped DNA and substrate, dCTP) forms; these different states have long been used to deduce that a large-scale conformational change may help the polymerase choose the correct nucleotide, and hence monitor DNA synthesis fidelity, through an "induced-fit" mechanism. However, the existence of open states with bound substrate and closed states without substrates suggest that substrate-induced conformational closing may be more subtle. Our dynamics simulations of two pol-beta/DNA systems (with/without substrates at the active site) reveal the large-scale closing motions of the thumb and 8-kDa subdomains in the presence of the correct substrate--leading to nearly perfect rearrangement of residues in the active site for the subsequent chemical step of nucleotidyl transfer--in contrast to an opening trend when the substrate is absent, leading to complete disassembly of the active site residues. These studies thus provide in silico evidence for the substrate-induced conformational rearrangements, as widely assumed based on a variety of crystallographic open and closed complexes. Further details gleaned from essential dynamics analyses clarify functionally relevant global motions of the polymerase-beta/DNA complex as required to prepare the system for the chemical reaction of nucleotide extension.
Figures
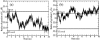



Similar articles
-
Differing conformational pathways before and after chemistry for insertion of dATP versus dCTP opposite 8-oxoG in DNA polymerase beta.Biophys J. 2007 May 1;92(9):3063-70. doi: 10.1529/biophysj.106.092106. Epub 2007 Feb 9. Biophys J. 2007. PMID: 17293403 Free PMC article.
-
Mismatch-induced conformational distortions in polymerase beta support an induced-fit mechanism for fidelity.Biochemistry. 2005 Oct 11;44(40):13328-41. doi: 10.1021/bi0507682. Biochemistry. 2005. PMID: 16201758
-
Sequential side-chain residue motions transform the binary into the ternary state of DNA polymerase lambda.Biophys J. 2006 Nov 1;91(9):3182-95. doi: 10.1529/biophysj.106.092080. Epub 2006 Aug 18. Biophys J. 2006. PMID: 16920835 Free PMC article.
-
Regulation of DNA repair fidelity by molecular checkpoints: "gates" in DNA polymerase beta's substrate selection.Biochemistry. 2006 Dec 26;45(51):15142-56. doi: 10.1021/bi061353z. Epub 2006 Dec 1. Biochemistry. 2006. PMID: 17176036 Free PMC article. Review.
-
Fidelity mechanisms of DNA polymerase beta.Prog Nucleic Acid Res Mol Biol. 2003;73:137-69. doi: 10.1016/s0079-6603(03)01005-5. Prog Nucleic Acid Res Mol Biol. 2003. PMID: 12882517 Review.
Cited by
-
Conformational coupling, bridge helix dynamics and active site dehydration in catalysis by RNA polymerase.Biochim Biophys Acta. 2010 Aug;1799(8):575-87. doi: 10.1016/j.bbagrm.2010.05.002. Epub 2010 May 15. Biochim Biophys Acta. 2010. PMID: 20478425 Free PMC article.
-
Insertion of oxidized nucleotide triggers rapid DNA polymerase opening.Nucleic Acids Res. 2016 May 19;44(9):4409-24. doi: 10.1093/nar/gkw174. Epub 2016 Mar 31. Nucleic Acids Res. 2016. PMID: 27034465 Free PMC article.
-
Effect of oxidatively damaged DNA on the active site preorganization during nucleotide incorporation in a high fidelity polymerase from Bacillus stearothermophilus.Proteins. 2008 May 15;71(3):1360-72. doi: 10.1002/prot.21824. Proteins. 2008. PMID: 18058909 Free PMC article.
-
Relationship between conformational changes in pol lambda's active site upon binding incorrect nucleotides and mismatch incorporation rates.J Phys Chem B. 2009 Oct 1;113(39):13035-47. doi: 10.1021/jp903172x. J Phys Chem B. 2009. PMID: 19572669 Free PMC article.
-
Computational simulation strategies for analysis of multisubunit RNA polymerases.Chem Rev. 2013 Nov 13;113(11):8546-66. doi: 10.1021/cr400046x. Epub 2013 Aug 29. Chem Rev. 2013. PMID: 23987500 Free PMC article. Review. No abstract available.
References
-
- Ahn, J., B. G. Werneburg, and M.-D. Tsai. 1997. DNA polymerase β: structure-fidelity relationship from pre-steady-state kinetic analyses of all possible correct and incorrect base pairs for wild type and R283A mutant. Biochemistry. 36:1100–1107. - PubMed
-
- Amadei, A., A. B. M. Linssen, and H. J. C. Berendsen. 1993. Essential dynamics of proteins. Proteins. 17:412–425. - PubMed
-
- Arcangeli, C., A. R. Bizzari, and S. Cannistraro. 2000a. Concerted motions in copper plastocyanin and azurin: an essential dynamics study. Biophys. Chem. 90:45–56. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

