Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor
- PMID: 15509782
- PMCID: PMC525479
- DOI: 10.1128/MCB.24.22.9771-9785.2004
Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor
Abstract
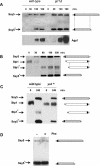
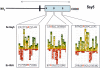
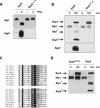
Saccharomyces cerevisiae cells possess a plasma membrane sensor able to detect the presence of extracellular amino acids and then to activate a signaling pathway leading to transcriptional induction of multiple genes, e.g., AGP1, encoding an amino acid permease. This sensing function requires the permease-like Ssy1 and associated Ptr3 and Ssy5 proteins, all essential to activation, by endoproteolytic processing, of the membrane-bound Stp1 transcription factor. The SCF(Grr1) ubiquitin-ligase complex is also essential to AGP1 induction, but its exact role in the amino acid signaling pathway remains unclear. Here we show that Stp1 undergoes casein kinase I-dependent phosphorylation. In the yck mutant lacking this kinase, Stp1 is not cleaved and AGP1 is not induced in response to amino acids. Furthermore, we provide evidence that Ssy5 is the endoprotease responsible for Stp1 processing. Ssy5 is significantly similar to serine proteases, its self-processing is a prerequisite for Stp1 cleavage, and its overexpression causes inducer-independent Stp1 cleavage and high-level AGP1 transcription. We further show that Stp1 processing also requires the SCF(Grr1) complex but is insensitive to proteasome inhibition. However, Stp1 processing does not require SCF(Grr1), Ssy1, or Ptr3 when Ssy5 is overproduced. Finally, we describe the properties of a particular ptr3 mutant that suggest that Ptr3 acts with Ssy1 in amino acid detection and signal initiation. We propose that Ssy1 and Ptr3 form the core components of the amino acid sensor. Upon detection of external amino acids, Ssy1-Ptr3 likely allows-in a manner dependent on SCF(Grr1)-the Ssy5 endoprotease to gain access to and to cleave Stp1, this requiring prior phosphorylation of Stp1 by casein kinase I.
Figures






References
-
- André, B., C. Hein, M. Grenson, and J. C. Jauniaux. 1993. Cloning and expression of the UGA4 gene coding for the inducible GABA-specific transport protein of Saccharomyces cerevisiae. Mol. Gen. Genet. 237:17-25. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
-
- Babu, P., J. D. Bryan, H. R. Panek, S. L. Jordan, B. M. Forbrich, S. C. Kelley, R. T. Colvin, and L. C. Robinson. 2002. Plasma membrane localization of the Yck2p yeast casein kinase 1 isoform requires the C-terminal extension and secretory pathway function. J. Cell Sci. 115:4957-4968. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
