Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA
- PMID: 15509786
- PMCID: PMC525493
- DOI: 10.1128/MCB.24.22.9823-9834.2004
Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA
Abstract
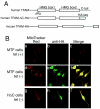
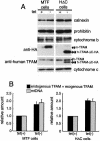
Mitochondrial transcription factor A (TFAM), a transcription factor for mitochondrial DNA (mtDNA) that also possesses the property of nonspecific DNA binding, is essential for maintenance of mtDNA. To clarify the role of TFAM, we repressed the expression of endogenous TFAM in HeLa cells by RNA interference. The amount of TFAM decreased maximally to about 15% of the normal level at day 3 after RNA interference and then recovered gradually. The amount of mtDNA changed closely in parallel with the daily change in TFAM while in organello transcription of mtDNA at day 3 was maintained at about 50% of the normal level. TFAM lacking its C-terminal 25 amino acids (TFAM-DeltaC) marginally activated transcription in vitro. When TFAM-DeltaC was expressed at levels comparable to those of endogenous TFAM in HeLa cells, mtDNA increased twofold, suggesting that TFAM-DeltaC is as competent in maintaining mtDNA as endogenous TFAM under these conditions. The in organello transcription of TFAM-DeltaC-expressing cells was no more than that in the control. Thus, the mtDNA amount is finely correlated with the amount of TFAM but not with the transcription level. We discuss an architectural role for TFAM in the maintenance of mtDNA in addition to its role in transcription activation.
Figures




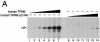
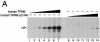
References
-
- Barat, M., D. Rickwood, C. Dufresne, and J. C. Mounolou. 1985. Characterization of DNA-protein complexes from the mitochondria of Xenopus laevis oocytes. Exp. Cell Res. 157:207-217. - PubMed
-
- Bowmaker, M., M. Y. Yang, T. Yasukawa, A. Reyes, H. T. Jacobs, J. A. Huberman, and I. J. Holt. 2003. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J. Biol. Chem. 278:50961-50969. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
