Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant
- PMID: 15510227
- PMCID: PMC524251
- DOI: 10.1371/journal.pbio.0020367
Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant
Abstract
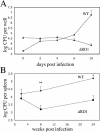
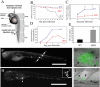
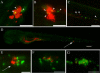
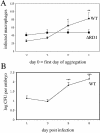
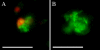
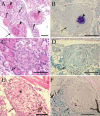
Granulomas are organized host immune structures composed of tightly interposed macrophages and other cells that form in response to a variety of persistent stimuli, both infectious and noninfectious. The tuberculous granuloma is essential for host containment of mycobacterial infection, although it does not always eradicate it. Therefore, it is considered a host-beneficial, if incompletely efficacious, immune response. The Mycobacterium RD1 locus encodes a specialized secretion system that promotes mycobacterial virulence by an unknown mechanism. Using transparent zebrafish embryos to monitor the infection process in real time, we found that RD1-deficient bacteria fail to elicit efficient granuloma formation despite their ability to grow inside of infected macrophages. We showed that macrophages infected with virulent mycobacteria produce an RD1-dependent signal that directs macrophages to aggregate into granulomas. This Mycobacterium-induced macrophage aggregation in turn is tightly linked to intercellular bacterial dissemination and increased bacterial numbers. Thus, mycobacteria co-opt host granulomas for their virulence.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures



Similar articles
-
The role of the granuloma in expansion and dissemination of early tuberculous infection.Cell. 2009 Jan 9;136(1):37-49. doi: 10.1016/j.cell.2008.11.014. Cell. 2009. PMID: 19135887 Free PMC article.
-
Looking within the zebrafish to understand the tuberculous granuloma.Adv Exp Med Biol. 2013;783:251-66. doi: 10.1007/978-1-4614-6111-1_13. Adv Exp Med Biol. 2013. PMID: 23468113 Review.
-
The secret lives of the pathogenic mycobacteria.Annu Rev Microbiol. 2003;57:641-76. doi: 10.1146/annurev.micro.57.030502.091033. Annu Rev Microbiol. 2003. PMID: 14527294 Review.
-
Mycobacterium tuberculosis-Induced Polarization of Human Macrophage Orchestrates the Formation and Development of Tuberculous Granulomas In Vitro.PLoS One. 2015 Jun 19;10(6):e0129744. doi: 10.1371/journal.pone.0129744. eCollection 2015. PLoS One. 2015. PMID: 26091535 Free PMC article.
-
Trafficking of superinfecting Mycobacterium organisms into established granulomas occurs in mammals and is independent of the Erp and ESX-1 mycobacterial virulence loci.J Infect Dis. 2008 Dec 15;198(12):1851-5. doi: 10.1086/593175. J Infect Dis. 2008. PMID: 18983252 Free PMC article.
Cited by
-
Mycobacterium marinum phthiocerol dimycocerosates enhance macrophage phagosomal permeabilization and membrane damage.PLoS One. 2020 Jul 23;15(7):e0233252. doi: 10.1371/journal.pone.0233252. eCollection 2020. PLoS One. 2020. PMID: 32701962 Free PMC article.
-
MiR-155-regulated molecular network orchestrates cell fate in the innate and adaptive immune response to Mycobacterium tuberculosis.Proc Natl Acad Sci U S A. 2016 Oct 11;113(41):E6172-E6181. doi: 10.1073/pnas.1608255113. Epub 2016 Sep 28. Proc Natl Acad Sci U S A. 2016. PMID: 27681624 Free PMC article.
-
Genomics and the evolution, pathogenesis, and diagnosis of tuberculosis.J Clin Invest. 2007 Jul;117(7):1738-45. doi: 10.1172/JCI31810. J Clin Invest. 2007. PMID: 17607348 Free PMC article. Review.
-
Exploring Macrophage-Dependent Wound Regeneration During Mycobacterial Infection in Zebrafish.Front Immunol. 2022 Mar 24;13:838425. doi: 10.3389/fimmu.2022.838425. eCollection 2022. Front Immunol. 2022. PMID: 35401552 Free PMC article.
-
EspM Is a Conserved Transcription Factor That Regulates Gene Expression in Response to the ESX-1 System.mBio. 2020 Feb 4;11(1):e02807-19. doi: 10.1128/mBio.02807-19. mBio. 2020. PMID: 32019792 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

