Oligo kernels for datamining on biological sequences: a case study on prokaryotic translation initiation sites
- PMID: 15511290
- PMCID: PMC535353
- DOI: 10.1186/1471-2105-5-169
Oligo kernels for datamining on biological sequences: a case study on prokaryotic translation initiation sites
Abstract
Background: Kernel-based learning algorithms are among the most advanced machine learning methods and have been successfully applied to a variety of sequence classification tasks within the field of bioinformatics. Conventional kernels utilized so far do not provide an easy interpretation of the learnt representations in terms of positional and compositional variability of the underlying biological signals.
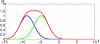
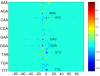
Results: We propose a kernel-based approach to datamining on biological sequences. With our method it is possible to model and analyze positional variability of oligomers of any length in a natural way. On one hand this is achieved by mapping the sequences to an intuitive but high-dimensional feature space, well-suited for interpretation of the learnt models. On the other hand, by means of the kernel trick we can provide a general learning algorithm for that high-dimensional representation because all required statistics can be computed without performing an explicit feature space mapping of the sequences. By introducing a kernel parameter that controls the degree of position-dependency, our feature space representation can be tailored to the characteristics of the biological problem at hand. A regularized learning scheme enables application even to biological problems for which only small sets of example sequences are available. Our approach includes a visualization method for transparent representation of characteristic sequence features. Thereby importance of features can be measured in terms of discriminative strength with respect to classification of the underlying sequences. To demonstrate and validate our concept on a biochemically well-defined case, we analyze E. coli translation initiation sites in order to show that we can find biologically relevant signals. For that case, our results clearly show that the Shine-Dalgarno sequence is the most important signal upstream a start codon. The variability in position and composition we found for that signal is in accordance with previous biological knowledge. We also find evidence for signals downstream of the start codon, previously introduced as transcriptional enhancers. These signals are mainly characterized by occurrences of adenine in a region of about 4 nucleotides next to the start codon.
Conclusions: We showed that the oligo kernel can provide a valuable tool for the analysis of relevant signals in biological sequences. In the case of translation initiation sites we could clearly deduce the most discriminative motifs and their positional variation from example sequences. Attractive features of our approach are its flexibility with respect to oligomer length and position conservation. By means of these two parameters oligo kernels can easily be adapted to different biological problems.
Figures
References
-
- Durbin R, Eddy SR, Krogh A. Biological Sequence Analysis. Cambridge University Press; 1998.
-
- Baldi P, Brunak S. Bioinformatics – The machine learning approach. Massachusetts Institute of Technology Press; 1998.
-
- Christiani N, Shawe-Taylor J. An Introduction to Support Vector Machines and other kernel-based learning methods. Cambridge University Press; 2000.
-
- Tikhonov AN, Arsenin VY. Solutions of ill-posed problems. Washington, DC: Winston; 1977.
-
- Degroeve S, Beats BD, de Peer YV, Rouzé P. Feature subset selection for splice site prediction. Bioinformatics. 2002;18:75–83. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

