d-matrix - database exploration, visualization and analysis
- PMID: 15511298
- PMCID: PMC533865
- DOI: 10.1186/1471-2105-5-168
d-matrix - database exploration, visualization and analysis
Abstract
Background: Motivated by a biomedical database set up by our group, we aimed to develop a generic database front-end with embedded knowledge discovery and analysis features. A major focus was the human-oriented representation of the data and the enabling of a closed circle of data query, exploration, visualization and analysis.
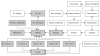
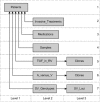
Results: We introduce a non-task-specific database front-end with a new visualization strategy and built-in analysis features, so called d-matrix. d-matrix is web-based and compatible with a broad range of database management systems. The graphical outcome consists of boxes whose colors show the quality of the underlying information and, as the name suggests, they are arranged in matrices. The granularity of the data display allows consequent drill-down. Furthermore, d-matrix offers context-sensitive categorization, hierarchical sorting and statistical analysis.
Conclusions: d-matrix enables data mining, with a high level of interactivity between humans and computer as a primary factor. We believe that the presented strategy can be very effective in general and especially useful for the integration of distinct data types such as phenotypical and molecular data.
Figures



References
-
- Genome Web http://www.hgmp.mrc.ac.uk/GenomeWeb/
-
- Kaynak B, von Heydebreck A, Mebus S, Seelow D, Hennig S, Vogel J, Sperling HP, Pregla R, Alexi-Meskishvili V, Hetzer R, Lange PE, Vingron M, Lehrach H, Sperling S. Genome-wide array analysis of normal and malformed human hearts. Circulation. 2003;107:2467–2474. doi: 10.1161/01.CIR.0000066694.21510.E2. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

