Mitotic checkpoint function in the formation of gross chromosomal rearrangements in Saccharomyces cerevisiae
- PMID: 15514023
- PMCID: PMC528767
- DOI: 10.1073/pnas.0407010101
Mitotic checkpoint function in the formation of gross chromosomal rearrangements in Saccharomyces cerevisiae
Abstract
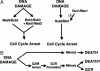
The accumulation of gross chromosomal rearrangements (GCRs) is characteristic of cancer cells. Multiple pathways that prevent GCRs, including S-phase cell cycle checkpoints, homologous recombination, telomere maintenance, suppression of de novo telomere addition, chromatin assembly, and mismatch repair, have been identified in Saccharomyces cerevisiae. However, pathways that promote the formation of GCRs are not as well understood. Of these, the de novo telomere addition pathway and nonhomologous end-joining are the best characterized. Here, we demonstrate that defects in the mitotic checkpoint and the mitotic exit network can suppress GCRs in strains containing defects that increase the GCR rate. These data suggest that functional mitotic checkpoints can play a role in the formation of genome rearrangements.
Figures

Similar articles
-
DNA repair pathway selection caused by defects in TEL1, SAE2, and de novo telomere addition generates specific chromosomal rearrangement signatures.PLoS Genet. 2014 Apr 3;10(4):e1004277. doi: 10.1371/journal.pgen.1004277. eCollection 2014 Apr. PLoS Genet. 2014. PMID: 24699249 Free PMC article.
-
Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2004 Jun 15;101(24):9039-44. doi: 10.1073/pnas.0403093101. Epub 2004 Jun 7. Proc Natl Acad Sci U S A. 2004. PMID: 15184655 Free PMC article.
-
Revisiting the role of the spindle assembly checkpoint in the formation of gross chromosomal rearrangements in Saccharomyces cerevisiae.Genetics. 2024 Nov 6;228(3):iyae150. doi: 10.1093/genetics/iyae150. Genetics. 2024. PMID: 39268895 Free PMC article.
-
Pathways and Mechanisms that Prevent Genome Instability in Saccharomyces cerevisiae.Genetics. 2017 Jul;206(3):1187-1225. doi: 10.1534/genetics.112.145805. Genetics. 2017. PMID: 28684602 Free PMC article. Review.
-
Measuring the rate of gross chromosomal rearrangements in Saccharomyces cerevisiae: A practical approach to study genomic rearrangements observed in cancer.Methods. 2007 Feb;41(2):168-76. doi: 10.1016/j.ymeth.2006.07.025. Methods. 2007. PMID: 17189859 Review.
Cited by
-
The Rad1-Rad10 complex promotes the production of gross chromosomal rearrangements from spontaneous DNA damage in Saccharomyces cerevisiae.Genetics. 2005 Apr;169(4):1927-37. doi: 10.1534/genetics.104.039768. Epub 2005 Jan 31. Genetics. 2005. PMID: 15687264 Free PMC article.
-
Inhibition of the mitotic exit network in response to damaged telomeres.PLoS Genet. 2013;9(10):e1003859. doi: 10.1371/journal.pgen.1003859. Epub 2013 Oct 10. PLoS Genet. 2013. PMID: 24130507 Free PMC article.
-
Esc2 orchestrates substrate-specific sumoylation by acting as a SUMO E2 cofactor in genome maintenance.Genes Dev. 2021 Feb 1;35(3-4):261-272. doi: 10.1101/gad.344739.120. Epub 2021 Jan 14. Genes Dev. 2021. PMID: 33446573 Free PMC article.
-
Sumoylation of the DNA polymerase ε by the Smc5/6 complex contributes to DNA replication.PLoS Genet. 2019 Nov 25;15(11):e1008426. doi: 10.1371/journal.pgen.1008426. eCollection 2019 Nov. PLoS Genet. 2019. PMID: 31765372 Free PMC article.
-
Living in CIN: Mitotic Infidelity and Its Consequences for Tumor Promotion and Suppression.Dev Cell. 2016 Dec 19;39(6):638-652. doi: 10.1016/j.devcel.2016.10.023. Dev Cell. 2016. PMID: 27997823 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

