Development of an expressed sequence tag (EST) resource for wheat (Triticum aestivum L.): EST generation, unigene analysis, probe selection and bioinformatics for a 16,000-locus bin-delineated map
- PMID: 15514037
- PMCID: PMC1448819
- DOI: 10.1534/genetics.104.034777
Development of an expressed sequence tag (EST) resource for wheat (Triticum aestivum L.): EST generation, unigene analysis, probe selection and bioinformatics for a 16,000-locus bin-delineated map
Abstract
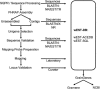
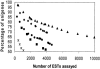
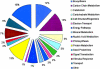
This report describes the rationale, approaches, organization, and resource development leading to a large-scale deletion bin map of the hexaploid (2n = 6x = 42) wheat genome (Triticum aestivum L.). Accompanying reports in this issue detail results from chromosome bin-mapping of expressed sequence tags (ESTs) representing genes onto the seven homoeologous chromosome groups and a global analysis of the entire mapped wheat EST data set. Among the resources developed were the first extensive public wheat EST collection (113,220 ESTs). Described are protocols for sequencing, sequence processing, EST nomenclature, and the assembly of ESTs into contigs. These contigs plus singletons (unassembled ESTs) were used for selection of distinct sequence motif unigenes. Selected ESTs were rearrayed, validated by 5' and 3' sequencing, and amplified for probing a series of wheat aneuploid and deletion stocks. Images and data for all Southern hybridizations were deposited in databases and were used by the coordinators for each of the seven homoeologous chromosome groups to validate the mapping results. Results from this project have established the foundation for future developments in wheat genomics.
Figures

References
-
- Adams, M. D., J. M. Kelley, J. D. Gocayne, M. Dubnick, M. H. Polymeropoulos et al., 1991. Complementary DNA sequencing: expressed sequence tags and human genome project. Science 252: 1651–1656. - PubMed

