Genetic dissection of nutritional copper signaling in chlamydomonas distinguishes regulatory and target genes
- PMID: 15514054
- PMCID: PMC1448816
- DOI: 10.1534/genetics.104.030460
Genetic dissection of nutritional copper signaling in chlamydomonas distinguishes regulatory and target genes
Abstract
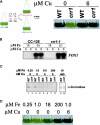
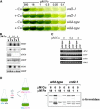
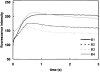
A genetic screen for Chlamydomonas reinhardtii mutants with copper-dependent growth or nonphotosynthetic phenotypes revealed three loci, COPPER RESPONSE REGULATOR 1 (CRR1), COPPER RESPONSE DEFECT 1 (CRD1), and COPPER RESPONSE DEFECT 2 (CRD2), distinguished as regulatory or target genes on the basis of phenotype. CRR1 was shown previously to be required for transcriptional activation of target genes like CYC6, CPX1, and CRD1, encoding, respectively, cytochrome c(6) (which is a heme-containing substitute for copper-containing plastocyanin), coproporphyrinogen III oxidase, and Mg-protoporphyrin IX monomethylester cyclase. We show here that CRR1 is required also for normal accumulation of copper proteins like plastocyanin and ferroxidase in copper-replete medium and for apoplastocyanin degradation in copper-deficient medium, indicating that a single pathway controls nutritional copper homeostasis at multiple levels. CRR1 is linked to the SUPPRESSOR OF PCY1-AC208 13 (SOP13) locus, which corresponds to a gain-of-function mutation resulting in copper-independent expression of CYC6. CRR1 is required also for hypoxic growth, pointing to a physiologically meaningful regulatory connection between copper deficiency and hypoxia. The growth phenotype of crr1 strains results primarily from secondary iron deficiency owing to reduced ferroxidase abundance, suggesting a role for CRR1 in copper distribution to a multicopper ferroxidase involved in iron assimilation. Mutations at the CRD2 locus also result in copper-conditional iron deficiency, which is consistent with a function for CRD2 in a pathway for copper delivery to the ferroxidase. Taken together, the observations argue for a specialized copper-deficiency adaptation for iron uptake in Chlamydomonas.
Figures





References
-
- Andrew, T. L., G. R. Pascale and H. A. Dailey, 1990 Regulation of heme biosynthesis in higher animals, pp. 163–200 in Biosynthesis of Heme and Chlorophylls, edited by H. A. Dailey. McGraw-Hill, New York.
-
- Askwith, C., and J. Kaplan, 1998. Iron and copper transport in yeast and its relevance to human disease. Trends Biochem. Sci. 23: 135–138. - PubMed
-
- Askwith, C., D. Eide, A. Van Ho, P. S. Bernard, L. Li et al., 1994. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76: 403–410. - PubMed
-
- Bull, P. C., G. R. Thomas, J. M. Rommens, J. R. Forbes and D. W. Cox, 1993. The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat. Genet. 5: 327–337. - PubMed
-
- Chelly, J., Z. Tumer, T. Tonnesen, A. Petterson, Y. Ishikawa-Brush et al., 1993. Isolation of a candidate gene for Menkes disease that encodes a potential heavy metal binding protein. Nat. Genet. 3: 14–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

