Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium
- PMID: 15514110
- PMCID: PMC528795
- DOI: 10.1093/nar/gkh910
Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium
Abstract
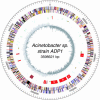
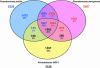
Acinetobacter sp. strain ADP1 is a nutritionally versatile soil bacterium closely related to representatives of the well-characterized Pseudomonas aeruginosa and Pseudomonas putida. Unlike these bacteria, the Acinetobacter ADP1 is highly competent for natural transformation which affords extraordinary convenience for genetic manipulation. The circular chromosome of the Acinetobacter ADP1, presented here, encodes 3325 predicted coding sequences, of which 60% have been classified based on sequence similarity to other documented proteins. The close evolutionary proximity of Acinetobacter and Pseudomonas species, as judged by the sequences of their 16S RNA genes and by the highest level of bidirectional best hits, contrasts with the extensive divergence in the GC content of their DNA (40 versus 62%). The chromosomes also differ significantly in size, with the Acinetobacter ADP1 chromosome <60% of the length of the Pseudomonas counterparts. Genome analysis of the Acinetobacter ADP1 revealed genes for metabolic pathways involved in utilization of a large variety of compounds. Almost all of these genes, with orthologs that are scattered in other species, are located in five major 'islands of catabolic diversity', now an apparent 'archipelago of catabolic diversity', within one-quarter of the overall genome. Acinetobacter ADP1 displays many features of other aerobic soil bacteria with metabolism oriented toward the degradation of organic compounds found in their natural habitat. A distinguishing feature of this genome is the absence of a gene corresponding to pyruvate kinase, the enzyme that generally catalyzes the terminal step in conversion of carbohydrates to pyruvate for respiration by the citric acid cycle. This finding supports the view that the cycle itself is centrally geared to the catabolic capabilities of this exceptionally versatile organism.
Figures

References
-
- Stanier R.Y., Palleroni,N.J. and Doudoroff,M. (1966) The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol., 43, 159–271. - PubMed
-
- Abdel-El-Haleem D. (2003) Acintobacter: environmental and biotechnological applications. Afr. J. Biotechnol., 2, 71–74.
-
- Smolyakov R., Borer,A., Riesenberg,K., Schlaeffer,F., Alkan,M., Porath,A., Rimar,D., Almog,Y. and Gilad,J. (2003) Nosocomial multi-drug resistant Acinetobacter baumannii bloodstream infection: risk factors and outcome with ampicillin-sulbactam treatment. J. Hosp. Infect., 54, 32–38. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

