Comparison of microsatellites versus single-nucleotide polymorphisms in a genome linkage screen for prostate cancer-susceptibility Loci
- PMID: 15514889
- PMCID: PMC1182157
- DOI: 10.1086/425870
Comparison of microsatellites versus single-nucleotide polymorphisms in a genome linkage screen for prostate cancer-susceptibility Loci
Abstract
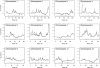
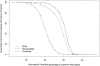
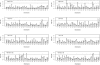
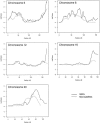
Prostate cancer is one of the most common cancers among men and has long been recognized to occur in familial clusters. Brothers and sons of affected men have a 2-3-fold increased risk of developing prostate cancer. However, identification of genetic susceptibility loci for prostate cancer has been extremely difficult. Although the suggestion of linkage has been reported for many chromosomes, the most promising regions have been difficult to replicate. In this study, we compare genome linkage scans using microsatellites with those using single-nucleotide polymorphisms (SNPs), performed in 467 men with prostate cancer from 167 families. For the microsatellites, the ABI Prism Linkage Mapping Set version 2, with 402 microsatellite markers, was used, and, for the SNPs, the Early Access Affymetrix Mapping 10K array was used. Our results show that the presence of linkage disequilibrium (LD) among SNPs can lead to inflated LOD scores, and this seems to be an artifact due to the assumption of linkage equilibrium that is required by the current genetic-linkage software. After excluding SNPs with high LD, we found a number of new LOD-score peaks with values of at least 2.0 that were not found by the microsatellite markers: chromosome 8, with a maximum model-free LOD score of 2.2; chromosome 2, with a LOD score of 2.1; chromosome 6, with a LOD score of 4.2; and chromosome 12, with a LOD score of 3.9. The LOD scores for chromosomes 6 and 12 are difficult to interpret, because they occurred only at the extreme ends of the chromosomes. The greatest gain provided by the SNP markers was a large increase in the linkage information content, with an average information content of 61% for the SNPs, versus an average of 41% for the microsatellite markers. The strengths and weaknesses of microsatellite versus SNP markers are illustrated by the results of our genome linkage scans.
Figures





References
Electronic-Database Information
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for prostate cancer, HPC1, PCAP, CAPB, HPC2, HPC20, and HPCX)
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

