Genetic inheritance of gene expression in human cell lines
- PMID: 15514893
- PMCID: PMC1182144
- DOI: 10.1086/426461
Genetic inheritance of gene expression in human cell lines
Abstract
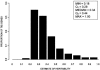
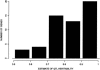
Combining genetic inheritance information, for both molecular profiles and complex traits, is a promising strategy not only for detecting quantitative trait loci (QTLs) for complex traits but for understanding which genes, pathways, and biological processes are also under the influence of a given QTL. As a primary step in determining the feasibility of such an approach in humans, we present the largest survey to date, to our knowledge, of the heritability of gene-expression traits in segregating human populations. In particular, we measured expression for 23,499 genes in lymphoblastoid cell lines for members of 15 Centre d'Etude du Polymorphisme Humain (CEPH) families. Of the total set of genes, 2,340 were found to be expressed, of which 31% had significant heritability when a false-discovery rate of 0.05 was used. QTLs were detected for 33 genes on the basis of at least one P value <.000005. Of these, 13 genes possessed a QTL within 5 Mb of their physical location. Hierarchical clustering was performed on the basis of both Pearson correlation of gene expression and genetic correlation. Both reflected biologically relevant activity taking place in the lymphoblastoid cell lines, with greater coherency represented in Kyoto Encyclopedia of Genes and Genomes database (KEGG) pathways than in Gene Ontology database pathways. However, more pathway coherence was observed in KEGG pathways when clustering was based on genetic correlation than when clustering was based on Pearson correlation. As more expression data in segregating populations are generated, viewing clusters or networks based on genetic correlation measures and shared QTLs will offer potentially novel insights into the relationship among genes that may underlie complex traits.
Figures


References
Electronic-Database Information
-
- CEPH Genotype Database, http://www.cephb.fr/cephdb/
-
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/index.html
-
- Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/ (for expression data for the 167 individuals utilized in the present study [GEO accession number GSE1726])
-
- Gene Ontology Consortium, http://www.geneontology.org/ (for the GO database)
-
- Kyoto Encyclopedia of Genes and Genomes (KEGG) Genes Database, http://www.genome.ad.jp/kegg/genes.html
References
-
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

