Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice
- PMID: 15516498
- PMCID: PMC527165
- DOI: 10.1104/pp.104.052274
Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice
Abstract
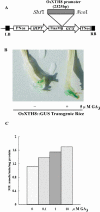
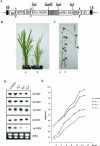
Xyloglucan endotransglucosylases/hydrolases (XTHs) that mediate cleavage and rejoining of the beta (1-4)-xyloglucans of the primary cell wall are considered to play an important role in the construction and restructuring of xyloglucan cross-links. A novel rice (Oryza sativa) XTH-related gene, OsXTH8, was cloned and characterized after being identified by cDNA microarray analysis of gibberellin-induced changes in gene expression in rice seedlings. OsXTH8 was a single copy gene; its full-length cDNA was 1,298 bp encoding a predicted protein of 290 amino acids. Phylogenetic analysis revealed that OsXTH8 falls outside of the three established subfamilies of XTH-related genes. OsXTH8 was preferentially expressed in rice leaf sheath in response to gibberellic acid. In situ hybridization and OsXTH8 promoter GUS fusion analysis revealed that OsXTH8 was highly expressed in vascular bundles of leaf sheath and young nodal roots where the cells are actively undergoing elongation and differentiation. OsXTH8 gene expression was up-regulated by gibberellic acid and there was very little effect of other hormones. In two genetic mutants of rice with abnormal height, the expression of OsXTH8 positively correlated with the height of the mutants. Transgenic rice expressing an RNAi construct of OsXTH8 exhibited repressed growth. These results indicate that OsXTH8 is differentially expressed in rice leaf sheath in relation to gibberellin and potentially involved in cell elongation processes.
Figures


References
-
- Arrowsmith DA, de Silva J (1995) Characterization of two tomato fruit-expressed cDNAs encoding xyloglucan endotransglucosylases. Plant Mol Biol 28: 391–403 - PubMed
-
- Barrachina C, Lorences EP (1998) Xyloglucan endotransglucosylase activity in pine hypocotyls: intracellular localization and relationship with endogenous growth. Physiol Plant 102: 55–60 - PubMed
-
- Borriss R, Buettner K, Maentsaelae P (1990) Structure of the β-1,3-1,4-glucanase gene of Bacillus macerans: homologies to other β-glucanases. Mol Gen Genet 222: 278–283 - PubMed
-
- Buckeridge MS, Rayon C, Urbanowics B, Tine Aurelio MAS, Carpita NC (2004) Mixed linkage (1–>3), (1–>4) β-D-glucans of grasses. Cereal Chem 18: 115–127
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

