AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis
- PMID: 15516508
- PMCID: PMC527164
- DOI: 10.1104/pp.104.047266
AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis
Abstract
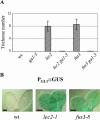
Embryonic regulators LEC2 (LEAFY COTYLEDON2) and FUS3 (FUSCA3) are involved in multiple aspects of Arabidopsis (Arabidopsis thaliana) seed development, including repression of leaf traits and premature germination and activation of seed storage protein genes. In this study, we show that gibberellin (GA) hormone biosynthesis is regulated by LEC2 and FUS3 pathways. The level of bioactive GAs is increased in immature seeds of lec2 and fus3 mutants relative to wild-type level. In addition, we show that the formation of ectopic trichome cells on lec2 and fus3 embryos is a GA-dependent process as in true leaves, suggesting that the GA pathway is misactivated in embryonic mutants. We next demonstrate that the GA-biosynthesis gene AtGA3ox2, which encodes the key enzyme AtGA3ox2 that catalyzes the conversion of inactive to bioactive GAs, is ectopically activated in embryos of the two mutants. Interestingly, both beta-glucuronidase reporter gene expression and in situ hybridization indicate that FUS3 represses AtGA3ox2 expression mainly in epidermal cells of embryo axis, which is distinct from AtGA3ox2 pattern at germination. Finally, we show that the FUS3 protein physically interacts with two RY elements (CATGCATG) present in the AtGA3ox2 promoter. This work suggests that GA biosynthesis is directly controlled by embryonic regulators during Arabidopsis embryonic development.
Figures



Similar articles
-
Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes.Plant Physiol. 2007 Feb;143(2):902-11. doi: 10.1104/pp.106.092320. Epub 2006 Dec 8. Plant Physiol. 2007. PMID: 17158584 Free PMC article.
-
FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana.New Phytol. 2017 Mar;213(4):1740-1754. doi: 10.1111/nph.14313. Epub 2016 Nov 23. New Phytol. 2017. PMID: 27878992
-
Regulation of storage protein gene expression in Arabidopsis.Development. 2003 Dec;130(24):6065-73. doi: 10.1242/dev.00814. Development. 2003. PMID: 14597573
-
LEAFY COTYLEDONs: old genes with new roles beyond seed development.F1000Res. 2019 Dec 27;8:F1000 Faculty Rev-2144. doi: 10.12688/f1000research.21180.1. eCollection 2019. F1000Res. 2019. PMID: 31942235 Free PMC article. Review.
-
Regulation of gibberellin biosynthesis by light.Curr Opin Plant Biol. 1999 Oct;2(5):398-403. doi: 10.1016/s1369-5266(99)00012-6. Curr Opin Plant Biol. 1999. PMID: 10508758 Review.
Cited by
-
The regulatory mechanism of chilling-induced dormancy transition from endo-dormancy to non-dormancy in Polygonatum kingianum Coll.et Hemsl rhizome bud.Plant Mol Biol. 2019 Feb;99(3):205-217. doi: 10.1007/s11103-018-0812-z. Epub 2019 Jan 9. Plant Mol Biol. 2019. PMID: 30627860
-
BASIC PENTACYSTEINE2 fine-tunes corm dormancy release in Gladiolus.Plant Physiol. 2023 Apr 3;191(4):2489-2505. doi: 10.1093/plphys/kiad026. Plant Physiol. 2023. PMID: 36659854 Free PMC article.
-
Understanding and Exploiting Post-Translational Modifications for Plant Disease Resistance.Biomolecules. 2021 Jul 30;11(8):1122. doi: 10.3390/biom11081122. Biomolecules. 2021. PMID: 34439788 Free PMC article. Review.
-
Voice from both sides: a molecular dialogue between transcriptional activators and repressors in seed-to-seedling transition and crop adaptation.Front Plant Sci. 2024 Aug 6;15:1416216. doi: 10.3389/fpls.2024.1416216. eCollection 2024. Front Plant Sci. 2024. PMID: 39166233 Free PMC article. Review.
-
The embryonic leaf identity gene FUSCA3 regulates vegetative phase transitions by negatively modulating ethylene-regulated gene expression in Arabidopsis.BMC Biol. 2012 Feb 20;10:8. doi: 10.1186/1741-7007-10-8. BMC Biol. 2012. PMID: 22348746 Free PMC article.
References
-
- Baumlein H, Misera S, Luerssen H, Kolle K, Horstmann C, Wobus U, Muller AJ (1994) The FUS3 gene of Arabidopsis thaliana is a regulator of gene expression during late embryogenesis. Plant J 6: 379–387
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

