Evolutionary dynamics of insertion sequences in Helicobacter pylori
- PMID: 15516562
- PMCID: PMC524885
- DOI: 10.1128/JB.186.22.7508-7520.2004
Evolutionary dynamics of insertion sequences in Helicobacter pylori
Abstract
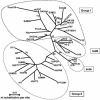
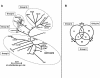
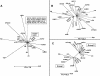
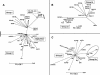
Prokaryotic insertion sequence (IS) elements behave like parasites in terms of their ability to invade and proliferate in microbial gene pools and like symbionts when they coevolve with their bacterial hosts. Here we investigated the evolutionary history of IS605 and IS607 of Helicobacter pylori, a genetically diverse gastric pathogen. These elements contain unrelated transposase genes (orfA) and also a homolog of the Salmonella virulence gene gipA (orfB). A total of 488 East Asian, Indian, Peruvian, and Spanish isolates were screened, and 18 and 14% of them harbored IS605 and IS607, respectively. IS605 nucleotide sequence analysis (n = 42) revealed geographic subdivisions similar to those of H. pylori; the geographic subdivision was blurred, however, due in part to homologous recombination, as indicated by split decomposition and homoplasy tests (homoplasy ratio, 0.56). In contrast, the IS607 populations (n = 44) showed strong geographic subdivisions with less homologous recombination (homoplasy ratio, 0.2). Diversifying selection (ratio of nonsynonymous change to synonymous change, >>1) was evident in approximately 15% of the IS605 orfA codons analyzed but not in the IS607 orfA codons. Diversifying selection was also evident in approximately 2% of the IS605 orfB and approximately 10% of the IS607 orfB codons analyzed. We suggest that the evolution of these elements reflects selection for optimal transposition activity in the case of IS605 orfA and for interactions between the OrfB proteins and other cellular constituents that potentially contribute to bacterial fitness. Taken together, similarities in IS elements and H. pylori population genetic structures and evidence of adaptive evolution in IS elements suggest that there is coevolution between these elements and their bacterial hosts.
Figures
References
-
- Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. - PubMed
-
- Bandelt, H. J., and A. W. Dress. 1992. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol. Phylogenet. Evol. 1:242-252. - PubMed
-
- Berg, D. E., and M. M. Howe. 1989. Mobile DNA. ASM Press, Washington, D.C.
-
- Bergelson, J., M. Kreitman, E. A. Stahl, and D. Tian. 2001. Evolutionary dynamics of plant R-genes. Science 292:2281-2285. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

