Sequence organization and insertion specificity of the novel chimeric ISHp609 transposable element of Helicobacter pylori
- PMID: 15516563
- PMCID: PMC524915
- DOI: 10.1128/JB.186.22.7521-7528.2004
Sequence organization and insertion specificity of the novel chimeric ISHp609 transposable element of Helicobacter pylori
Abstract
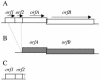
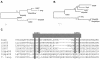
Here we describe ISHp609 of Helicobacter pylori, a new member of the IS605 mobile element family that is novel and contains two genes whose functions are unknown, jhp960 and jhp961, in addition to homologs of two other H. pylori insertion sequence (IS) element genes, orfA, which encodes a putative serine recombinase-transposase, and orfB, whose homologs in other species are also often annotated as genes that encode transposases. The complete four-gene element was found in 10 to 40% of strains obtained from Africa, India, Europe, and the Americas but in only 1% of East Asian strains. Sequence comparison of 10 representative ISHp609 elements revealed higher levels of DNA sequence matches (99%) than those seen in normal chromosomal genes (88 to 98%) or in other IS elements (95 to 97% for IS605, IS606, and IS607) from the same H. pylori populations. Sequence analysis suggested that ISHp609 can insert at many genomic sites with its left end preferentially next to TAT, with no target specificity for its right end, and without duplicating or deleting target sequences. A deleted form of ISHp609, containing just jhp960 and jhp961 and 37 bp of orfA, found in reference strain J99, was at the same chromosomal site in 15 to 40% of the strains from many geographic regions but again in only 1% of the East Asian strains. The abundance and sequence homogeneity of ISHp609 and of this nonmobile remnant suggested a recent bottleneck and then rapid spread in H. pylori populations, possibly selected by the contributions of the elements to bacterial fitness.
Figures

Similar articles
-
Functional organization and insertion specificity of IS607, a chimeric element of Helicobacter pylori.J Bacteriol. 2000 Oct;182(19):5300-8. doi: 10.1128/JB.182.19.5300-5308.2000. J Bacteriol. 2000. PMID: 10986230 Free PMC article.
-
Evolutionary dynamics of insertion sequences in Helicobacter pylori.J Bacteriol. 2004 Nov;186(22):7508-20. doi: 10.1128/JB.186.22.7508-7520.2004. J Bacteriol. 2004. PMID: 15516562 Free PMC article.
-
Transposable element ISHp608 of Helicobacter pylori: nonrandom geographic distribution, functional organization, and insertion specificity.J Bacteriol. 2002 Feb;184(4):992-1002. doi: 10.1128/jb.184.4.992-1002.2002. J Bacteriol. 2002. PMID: 11807059 Free PMC article.
-
Contributions of genome sequencing to understanding the biology of Helicobacter pylori.Annu Rev Microbiol. 1999;53:353-87. doi: 10.1146/annurev.micro.53.1.353. Annu Rev Microbiol. 1999. PMID: 10547695 Review.
-
The IS200/IS605 Family and "Peel and Paste" Single-strand Transposition Mechanism.Microbiol Spectr. 2015 Aug;3(4). doi: 10.1128/microbiolspec.MDNA3-0039-2014. Microbiol Spectr. 2015. PMID: 26350330 Review.
Cited by
-
Genetic Markers of Genome Rearrangements in Helicobacter pylori.Microorganisms. 2021 Mar 17;9(3):621. doi: 10.3390/microorganisms9030621. Microorganisms. 2021. PMID: 33802974 Free PMC article.
-
Helicobacter Pylori's plasticity zones are novel transposable elements.PLoS One. 2009 Sep 3;4(9):e6859. doi: 10.1371/journal.pone.0006859. PLoS One. 2009. PMID: 19727398 Free PMC article.
-
Insertion sequence content reflects genome plasticity in strains of the root nodule actinobacterium Frankia.BMC Genomics. 2009 Oct 12;10:468. doi: 10.1186/1471-2164-10-468. BMC Genomics. 2009. PMID: 19821988 Free PMC article.
-
The detection of inherent homologous recombination between repeat sequences in H. pylori 26695 by the PCR-based method.Curr Microbiol. 2014 Feb;68(2):211-9. doi: 10.1007/s00284-013-0466-7. Epub 2013 Oct 6. Curr Microbiol. 2014. PMID: 24097137
-
Phylogenetic Relationships among TnpB-Containing Mobile Elements in Six Bacterial Species.Genes (Basel). 2023 Feb 19;14(2):523. doi: 10.3390/genes14020523. Genes (Basel). 2023. PMID: 36833450 Free PMC article.
References
-
- Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. - PubMed
-
- Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A., Smith, and K. A. Struhl. 1994. Current protocols in molecular biology, supplement 27 CPMB, page 2.4.1. Greene Publishing and Wiley Interscience, New York, N.Y.
-
- Berg, D. E., R. H. Gilman, J. Lelwala-Guruge, K. Srivastava, Y. Valdez, J. Watanabe, J. Miyagi, N. S. Akopyants, A. Ramirez-Ramos, T. H. Yoshiwara, S. Recavarren, and R. Leon-Barua. 1997. Helicobacter pylori populations in individual Peruvian patients. Clin. Infect. Dis. 25:996-1002. - PubMed
-
- Berg, D. E., and M. M. Howe (ed.). 1989. Mobile DNA. American Society for Microbiology, Washington, D.C.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

