Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon
- PMID: 15516564
- PMCID: PMC524886
- DOI: 10.1128/JB.186.22.7529-7537.2004
Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon
Abstract
The flhD operon is the master operon of the flagellar regulon and a global regulator of metabolism. The genome sequence of the Escherichia coli K-12 strain MG1655 contained an IS1 insertion sequence element in the regulatory region of the flhD promoter. Another stock of MG1655 was obtained from the E. coli Genetic Stock Center. This stock contained isolates which were poorly motile and had no IS1 element upstream of the flhD promoter. From these isolates, motile subpopulations were identified after extended incubation in motility agar. Purified motile derivatives contained an IS5 element insertion upstream of the flhD promoter, and swarm rates were sevenfold higher than that of the original isolate. For a motile derivative, levels of flhD transcript had increased 2.7-fold, leading to a 32-fold increase in fliA transcript and a 65-fold increase in flhB::luxCDABE expression from a promoter probe vector. A collection of commonly used lab strains was screened for IS element insertion and motility. Five strains (RP437, YK410, MC1000, W3110, and W2637) contained IS5 elements upstream of the flhD promoter at either of two locations. This correlated with high swarm rates. Four other strains (W1485, FB8, MM294, and RB791) did not contain IS elements in the flhD regulatory region and were poorly motile. Primer extension determined that the transcriptional start site of flhD was unaltered by the IS element insertions. We suggest that IS element insertion may activate transcription of the flhD operon by reducing transcriptional repression.
Figures
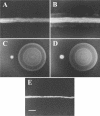
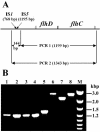


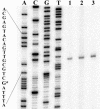
References
-
- Adler, J., and B. Templeton. 1967. The effect of environmental conditions on Escherichia coli. J. Gen. Microbiol. 46:175-184. - PubMed
-
- Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

