Molecular identification of the urea uptake system and transcriptional analysis of urea transporter- and urease-encoding genes in Corynebacterium glutamicum
- PMID: 15516578
- PMCID: PMC524917
- DOI: 10.1128/JB.186.22.7645-7652.2004
Molecular identification of the urea uptake system and transcriptional analysis of urea transporter- and urease-encoding genes in Corynebacterium glutamicum
Abstract
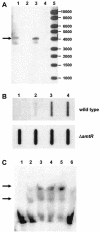
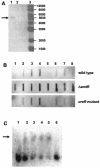
The molecular identification of the Corynebacterium glutamicum urea uptake system is described. This ABC-type transporter is encoded by the urtABCDE operon, which is transcribed in response to nitrogen limitation. Expression of the urt genes is regulated by the global nitrogen regulator AmtR, and an amtR deletion strain showed constitutive expression of the urtABCDE genes. The AmtR repressor protein also controls transcription of the urease-encoding ureABCEFGD genes in C. glutamicum. The ure gene cluster forms an operon which is mainly transcribed in response to nitrogen starvation. To confirm the increased synthesis of urease subunits under nitrogen limitation, proteome analyses of cytoplasmic protein extracts from cells grown under nitrogen surplus and nitrogen limitation were carried out, and five of the seven urease subunits were identified.
Figures

References
-
- Abe, S., K. Takayama, and S. Kinoshita. 1967. Taxonomical studies on glutamic acid-producing bacteria. J. Gen. Microbiol. 13:279-301.
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
-
- Beckers, G., L. Nolden, and A. Burkovski. 2001. Glutamate synthase of Corynebacterium glutamicum is not essential for glutamate synthesis and is regulated by the nitrogen status. Microbiology 147:2961-2970. - PubMed
-
- Burkovski, A. 2003. I do it my way: regulation of ammonium uptake and ammonium assimilation in Corynebacterium glutamicum. Arch. Microbiol. 179:83-88. - PubMed
-
- Burkovski, A. 2003. Ammonium assimilation and nitrogen control in Corynebacterium glutamicum and its relatives: an example for new regulatory mechanisms in actinomycetes. FEMS Microbiol. Rev. 27:617-628. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

