The bcr1 DNA repeat element is specific to the Bacillus cereus group and exhibits mobile element characteristics
- PMID: 15516586
- PMCID: PMC524882
- DOI: 10.1128/JB.186.22.7714-7725.2004
The bcr1 DNA repeat element is specific to the Bacillus cereus group and exhibits mobile element characteristics
Abstract
Bacillus cereus strains ATCC 10987 and ATCC 14579 harbor an approximately 155-bp repeated element, bcr1, which is conserved in B. cereus, B. anthracis, B. thuringiensis, and B. mycoides but not in B. subtilis and B. licheniformis. In this study, we show by Southern blot hybridizations that bcr1 is present in all 54 B. cereus group strains tested but absent in 11 Bacillus strains outside the group, suggesting that bcr1 may be specific and ubiquitous to the B. cereus group. By comparative analysis of the complete genome sequences of B. cereus ATCC 10987, B. cereus ATCC 14579, and B. anthracis Ames, we show that bcr1 is exclusively present in the chromosome but absent from large plasmids carried by these strains and that the numbers of full-length bcr1 repeats for these strains are 79, 54, and 12, respectively. Numerous copies of partial bcr1 elements are also present in the three genomes (91, 128, and 53, respectively). Furthermore, the genomic localization of bcr1 is not conserved between strains with respect to chromosomal position or organization of gene neighbors, as only six full-length bcr1 loci are common to at least two of the three strains. However, the intergenic sequence surrounding a specific bcr1 repeat in one of the three strains is generally strongly conserved in the other two, even in loci where bcr1 is found exclusively in one strain. This finding indicates that bcr1 either has evolved by differential deletion from a very high number of repeats in a common ancestor to the B. cereus group or is moving around the chromosome. The identification of bcr1 repeats interrupting genes in B. cereus ATCC 10987 and ATCC 14579 and the presence of a flanking TTTAT motif in each end show that bcr1 exhibits features characteristic of a mobile element.
Figures




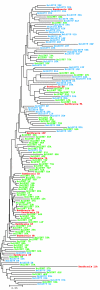


Similar articles
-
Interspersed DNA repeats bcr1-bcr18 of Bacillus cereus group bacteria form three distinct groups with different evolutionary and functional patterns.Mol Biol Evol. 2011 Feb;28(2):963-83. doi: 10.1093/molbev/msq269. Epub 2010 Oct 20. Mol Biol Evol. 2011. PMID: 20961964
-
Exploring the evolution of the Bacillus cereus group repeat element bcr1 by comparative genome analysis of closely related strains.Microbiology (Reading). 2007 Nov;153(Pt 11):3894-3908. doi: 10.1099/mic.0.2007/005504-0. Microbiology (Reading). 2007. PMID: 17975097
-
Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis.Nature. 2003 May 1;423(6935):87-91. doi: 10.1038/nature01582. Nature. 2003. PMID: 12721630
-
The Bacillus cereus group: novel aspects of population structure and genome dynamics.J Appl Microbiol. 2006 Sep;101(3):579-93. doi: 10.1111/j.1365-2672.2006.03087.x. J Appl Microbiol. 2006. PMID: 16907808 Review.
-
What sets Bacillus anthracis apart from other Bacillus species?Annu Rev Microbiol. 2009;63:451-76. doi: 10.1146/annurev.micro.091208.073255. Annu Rev Microbiol. 2009. PMID: 19514852 Review.
Cited by
-
Impact of small repeat sequences on bacterial genome evolution.Genome Biol Evol. 2011;3:959-73. doi: 10.1093/gbe/evr077. Epub 2011 Jul 29. Genome Biol Evol. 2011. PMID: 21803768 Free PMC article.
-
A combined de novo assembly approach increases the quality of prokaryotic draft genomes.Folia Microbiol (Praha). 2022 Oct;67(5):801-810. doi: 10.1007/s12223-022-00980-7. Epub 2022 Jun 6. Folia Microbiol (Praha). 2022. PMID: 35668290
-
Diversity of Bacillus cereus sensu lato mobilome.BMC Genomics. 2019 May 29;20(1):436. doi: 10.1186/s12864-019-5764-4. BMC Genomics. 2019. PMID: 31142281 Free PMC article.
-
Global mRNA decay analysis at single nucleotide resolution reveals segmental and positional degradation patterns in a Gram-positive bacterium.Genome Biol. 2012 Apr 26;13(4):R30. doi: 10.1186/gb-2012-13-4-r30. Genome Biol. 2012. PMID: 22537947 Free PMC article.
-
Detection of small RNAs in Bordetella pertussis and identification of a novel repeated genetic element.BMC Genomics. 2011 Apr 27;12:207. doi: 10.1186/1471-2164-12-207. BMC Genomics. 2011. PMID: 21524285 Free PMC article.
References
-
- Ash, C., J. A. Farrow, S. Wallbanks, and M. D. Collins. 1991. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett Appl. Microbiol. 13:202-206.
-
- Berry, C., S. O'Neil, E. Ben-Dov, A. F. Jones, L. Murphy, M. A. Quail, M. T. Holden, D. Harris, A. Zaritsky, and J. Parkhill. 2002. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 68:5082-5095. - PMC - PubMed
-
- Buisine, N., C. M. Tang, and R. Chalmers. 2002. Transposon-like Correia elements: structure, distribution and genetic exchange between pathogenic Neisseria sp. FEBS Lett. 522:52-58. - PubMed
-
- Chandler, M. 1998. Insertion sequences and transposons, p. 30-37. In F. J. De Bruijn, J. R Lupski, and G. Weinstock (ed.), Bacterial genomes—physical structure and analysis. Chapman & Hall, New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

