A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1
- PMID: 15520230
- PMCID: PMC2172501
- DOI: 10.1083/jcb.200405153
A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1
Abstract
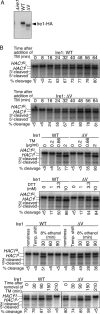
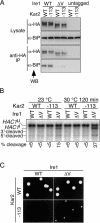
In the unfolded protein response, the type I transmembrane protein Ire1 transmits an endoplasmic reticulum (ER) stress signal to the cytoplasm. We previously reported that under nonstressed conditions, the ER chaperone BiP binds and represses Ire1. It is still unclear how this event contributes to the overall regulation of Ire1. The present Ire1 mutation study shows that the luminal domain possesses two subregions that seem indispensable for activity. The BiP-binding site was assigned not to these subregions, but to a region neighboring the transmembrane domain. Phenotypic comparison of several Ire1 mutants carrying deletions in the indispensable subregions suggests these subregions are responsible for multiple events that are prerequisites for activation of the overall Ire1 proteins. Unexpectedly, deletion of the BiP-binding site rendered Ire1 unaltered in ER stress inducibility, but hypersensitive to ethanol and high temperature. We conclude that in the ER stress-sensory system BiP is not the principal determinant of Ire1 activity, but an adjustor for sensitivity to various stresses.
Figures





References
-
- Bertolotti, A., Y. Zhang, L.M. Hendershot, H.P. Harding, and D. Ron. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326–332. - PubMed
-
- Calfon, M., H. Zeng, F. Urano, J.H. Till, S.R. Hubbard, H.P. Harding, S.G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 415:92–96. - PubMed
-
- Casagrande, R., P. Stern, M. Diehn, C. Shamu, M. Osario, M. Zuniga, P.O. Brown, and H. Ploegh. 2000. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell. 5:729–735. - PubMed
-
- Cox, J.S., and P. Walter. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 87:391–404. - PubMed
-
- Cox, J.S., C.E. Shamu, and P. Walter. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 73:1197–1206. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

