Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging
- PMID: 15520276
- PMCID: PMC528894
- DOI: 10.1101/gad.310304
Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging
Abstract
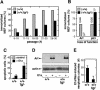
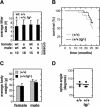
Mammalian genes frequently present allelic variants that differ in their expression levels and that, in the case of tumor suppressor genes, can be of relevance for cancer susceptibility and aging. We report here the characterization of a novel mouse model with increased activity for the Ink4a and Arf tumor suppressors. We have generated a "super Ink4a/Arf" mouse strain carrying a transgenic copy of the entire Ink4a/Arf locus. Cells derived from super Ink4a/Arf mice have increased resistance to in vitro immortalization and oncogenic transformation. Importantly, super Ink4a/Arf mice manifest higher resistance to cancer compared to normal, nontransgenic, mice. Finally, super Ink4a/Arf mice have normal aging and lifespan. Together, these results indicate that modest increases in the activity of the Ink4a/Arf tumor suppressor result in a beneficial cancer-resistant phenotype without affecting normal viability or aging.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
