Isolation of the repertoire of VSG expression site containing telomeres of Trypanosoma brucei 427 using transformation-associated recombination in yeast
- PMID: 15520294
- PMCID: PMC525691
- DOI: 10.1101/gr.2955304
Isolation of the repertoire of VSG expression site containing telomeres of Trypanosoma brucei 427 using transformation-associated recombination in yeast
Abstract
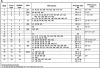
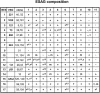
Trypanosoma brucei switches between variant surface glycoproteins (VSGs) allowing immune escape. The active VSG is in one of many telomeric bloodstream form VSG expression sites (BESs), also containing expression site-associated genes (ESAGs) involved in host adaptation. The role of BES sequence diversity in parasite virulence can best be understood through analysis of the full repertoire of BESs from a given T. brucei strain. However, few BESs have been cloned, as telomeres are highly underrepresented in standard libraries. We devised a strategy for isolating the repertoire of T. brucei 427 BES-containing telomeres in Saccaromyces cerevisiae by using transformation-associated recombination (TAR). We isolated 182 T. brucei 427 BES TAR clones, 167 of which could be subdivided into minimally 17 BES groups. This set gives us the first view of the breadth and diversity of BESs from one T. brucei strain. Most BESs ranged between 40 and 70 kb (average, 57 +/- 17 kb) and contained most identified ESAGs. Phylogenetic comparison of the cohort of BES promoter and ESAG6 sequences did not show similar trees, indicating rapid evolution most likely mediated by sequence exchange between BESs. This cloning strategy could be used for any T. brucei strain, facilitating research on the biodiversity of telomeric gene families and host-pathogen interactions.
Figures






References
-
- Ansorge, I., Steverding, D., Melville, S., Hartmann, C., and Clayton, C. 1999. Transcription of “inactive” expression sites in African trypanosomes leads to expression of multiple transferrin receptor RNAs in bloodstream forms. Mol. Biochem. Parasitol. 101: 81-94. - PubMed
-
- Ashcroft, M.T. 1958. The importance of African wild animals as reservoirs of trypanosomiasis. East African Med. J. 36: 289-297. - PubMed
-
- Barry, J.D. and McCulloch, R. 2001. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 49: 1-70. - PubMed
-
- Bernards, A., Van der Ploeg, L.H., Frasch, A.C., Borst, P., Boothroyd, J.C., Coleman, S., and Cross, G.A. 1981. Activation of trypanosome surface glycoprotein genes involves a duplication-transposition leading to an altered 3′ end. Cell 27: 497-505. - PubMed
-
- Berriman, M., Hall, N., Sheader, K., Bringaud, F., Tiwari, B., Isobe, T., Bowman, S., Corton, C., Clark, L., Cross, G.A., et al. 2002. The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 122: 131-140. - PubMed
WEB SITE REFERENCES
-
- http://bacpac.chori.org; BACPAC Resource Center with information about BAC libraries.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
