OptStrain: a computational framework for redesign of microbial production systems
- PMID: 15520298
- PMCID: PMC525696
- DOI: 10.1101/gr.2872004
OptStrain: a computational framework for redesign of microbial production systems
Abstract
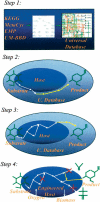
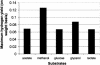
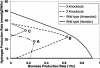
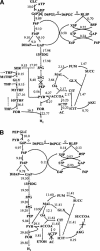
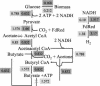
This paper introduces the hierarchical computational framework OptStrain aimed at guiding pathway modifications, through reaction additions and deletions, of microbial networks for the overproduction of targeted compounds. These compounds may range from electrons or hydrogen in biofuel cell and environmental applications to complex drug precursor molecules. A comprehensive database of biotransformations, referred to as the Universal database (with >5700 reactions), is compiled and regularly updated by downloading and curating reactions from multiple biopathway database sources. Combinatorial optimization is then used to elucidate the set(s) of non-native functionalities, extracted from this Universal database, to add to the examined production host for enabling the desired product formation. Subsequently, competing functionalities that divert flux away from the targeted product are identified and removed to ensure higher product yields coupled with growth. This work represents an advancement over earlier efforts by establishing an integrated computational framework capable of constructing stoichiometrically balanced pathways, imposing maximum product yield requirements, pinpointing the optimal substrate(s), and evaluating different microbial hosts. The range and utility of OptStrain are demonstrated by addressing two very different product molecules. The hydrogen case study pinpoints reaction elimination strategies for improving hydrogen yields using two different substrates for three separate production hosts. In contrast, the vanillin study primarily showcases which non-native pathways need to be added into Escherichia coli. In summary, OptStrain provides a useful tool to aid microbial strain design and, more importantly, it establishes an integrated framework to accommodate future modeling developments.
Figures


References
-
- Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, New York.
-
- Arita, M. 2000. Metabolic construction using shortest paths. Simulation Practice Theory 8: 109-125.
-
- Bailey, J.E. 2001. Complex biology with no parameters. Nat. Biotechnol. 19: 503-504. - PubMed
WEB SITE REFERENCES
-
- http://fenske.che.psu.edu/Faculty/CMaranas/pubs.html; Universal reaction database.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
