Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation
- PMID: 15522967
- PMCID: PMC524698
- DOI: 10.1073/pnas.0407044101
Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation
Abstract
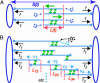
It has been proposed that the suppression of poleward flux within interpolar microtubule (ipMT) bundles of Drosophila embryonic spindles couples outward forces generated by a sliding filament mechanism to anaphase spindle elongation. Here, we (i) propose a molecular mechanism in which the bipolar kinesin KLP61F persistently slides dynamically unstable ipMTs outward, the MT depolymerase KLP10A acts at the poles to convert ipMT sliding to flux, and the chromokinesin KLP3A inhibits the depolymerase to suppress flux, thereby coupling ipMT sliding to spindle elongation; (ii) used KLP3A inhibitors to interfere with the coupling process, which revealed an inverse linear relation between the rates of flux and elongation, supporting the proposed mechanism and demonstrating that the suppression of flux controls both the rate and onset of spindle elongation; and (iii) developed a mathematical model using force balance and rate equations to describe how motors sliding the highly dynamic ipMTs apart can drive spindle elongation at a steady rate determined by the extent of suppression of flux.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

