Visualization of the movement of single histidine kinase molecules in live Caulobacter cells
- PMID: 15522969
- PMCID: PMC528753
- DOI: 10.1073/pnas.0404200101
Visualization of the movement of single histidine kinase molecules in live Caulobacter cells
Abstract
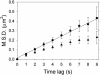
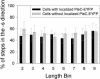
The bacterium Caulobacter crescentus divides asymmetrically as part of its normal life cycle. This asymmetry is regulated in part by the membrane-bound histidine kinase PleC, which localizes to one pole of the cell at specific times in the cell cycle. Here, we track single copies of PleC labeled with enhanced yellow fluorescent protein (EYFP) in the membrane of live Caulobacter cells over a time scale of seconds. In addition to the expected molecules immobilized at one cell pole, we observed molecules moving throughout the cell membrane. By tracking the positions of these molecules for several seconds, we determined a diffusion coefficient (D) of 12 +/- 2 x 10(-3) microm(2)/s for the mobile copies of PleC not bound at the cell pole. This D value is maintained across all cell cycle stages. We observe a reduced D at poles containing localized PleC-EYFP; otherwise D is independent of the position of the diffusing molecule within the bacterium. We did not detect any directional bias in the motion of the PleC-EYFP molecules, implying that the molecules are not being actively transported.
Figures

References
-
- Kadner, R. J. (1996) in Escherichia coli and Salmonella, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), Vol. 1, pp. 58–87.
-
- Miller, C. G. (1996) in Escherichia coli and Salmonella, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), Vol. 1, pp. 938–954.
-
- Jensen, R. B., Wang, S. C. & Shapiro, L. (2002) Nat. Rev. Mol. Cell. Biol. 3, 167–176. - PubMed
-
- Howard, M., Rutenberg, A. D. & de Vet, S. (2001) Phys. Rev. Lett 87, 278102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

