Folding of human superoxide dismutase: disulfide reduction prevents dimerization and produces marginally stable monomers
- PMID: 15522970
- PMCID: PMC528748
- DOI: 10.1073/pnas.0403979101
Folding of human superoxide dismutase: disulfide reduction prevents dimerization and produces marginally stable monomers
Abstract
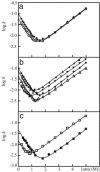
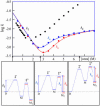
The molecular mechanism by which the homodimeric enzyme Cu/Zn superoxide dismutase (SOD) causes neural damage in amytrophic lateral sclerosis is yet poorly understood. A striking, as well as an unusual, feature of SOD is that it maintains intrasubunit disulfide bonds in the reducing environment of the cytosol. Here, we investigate the role of these disulfide bonds in folding and assembly of the SOD apo protein (apoSOD) homodimer through extensive protein engineering. The results show that apoSOD folds in a simple three-state process by means of two kinetic barriers: 2D<==>2M<==>M(2). The early predominant barrier represents folding of the monomers (M), and the late barrier the assembly of the dimer (M(2)). Unique for this mechanism is a dependence of protein concentration on the unfolding rate constant under physiological conditions, which disappears above 6 M Urea where the transition state for unfolding shifts to first-order dissociation of the dimer in accordance with Hammond-postulate behavior. Although reduction of the intrasubunit disulfide bond C57-C146 is not critical for folding of the apoSOD monomer, it has a pronounced effect on its stability and abolishes subsequent dimerization. Thus, impaired ability to form, or retain, the C57-C146 bond in vivo is predicted to increase the cellular load of marginally stable apoSOD monomers, which may have implications for the amytrophic lateral sclerosis neuropathology.
Figures
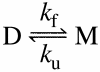
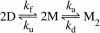
Similar articles
-
The rate and equilibrium constants for a multistep reaction sequence for the aggregation of superoxide dismutase in amyotrophic lateral sclerosis.Proc Natl Acad Sci U S A. 2004 Oct 19;101(42):15094-9. doi: 10.1073/pnas.0406650101. Epub 2004 Oct 8. Proc Natl Acad Sci U S A. 2004. PMID: 15475574 Free PMC article.
-
The coupling between disulphide status, metallation and dimer interface strength in Cu/Zn superoxide dismutase.J Mol Biol. 2007 Jan 12;365(2):333-42. doi: 10.1016/j.jmb.2006.09.048. Epub 2006 Sep 23. J Mol Biol. 2007. PMID: 17070542
-
The Disulfide Bond, but Not Zinc or Dimerization, Controls Initiation and Seeded Growth in Amyotrophic Lateral Sclerosis-linked Cu,Zn Superoxide Dismutase (SOD1) Fibrillation.J Biol Chem. 2015 Dec 18;290(51):30624-36. doi: 10.1074/jbc.M115.666503. Epub 2015 Oct 28. J Biol Chem. 2015. PMID: 26511321 Free PMC article.
-
Posttranslational modifications in Cu,Zn-superoxide dismutase and mutations associated with amyotrophic lateral sclerosis.Antioxid Redox Signal. 2006 May-Jun;8(5-6):847-67. doi: 10.1089/ars.2006.8.847. Antioxid Redox Signal. 2006. PMID: 16771675 Free PMC article. Review.
-
Structure, folding, and misfolding of Cu,Zn superoxide dismutase in amyotrophic lateral sclerosis.Biochim Biophys Acta. 2006 Nov-Dec;1762(11-12):1025-37. doi: 10.1016/j.bbadis.2006.05.004. Epub 2006 May 22. Biochim Biophys Acta. 2006. PMID: 16814528 Review.
Cited by
-
Fibrillation precursor of superoxide dismutase 1 revealed by gradual tuning of the protein-folding equilibrium.Proc Natl Acad Sci U S A. 2012 Oct 30;109(44):17868-73. doi: 10.1073/pnas.1201795109. Epub 2012 Jul 13. Proc Natl Acad Sci U S A. 2012. PMID: 22797895 Free PMC article.
-
Cu,Zn-superoxide dismutase increases toxicity of mutant and zinc-deficient superoxide dismutase by enhancing protein stability.J Biol Chem. 2010 Oct 29;285(44):33885-97. doi: 10.1074/jbc.M110.118901. Epub 2010 Jul 27. J Biol Chem. 2010. PMID: 20663894 Free PMC article.
-
Experimental Mutations in Superoxide Dismutase 1 Provide Insight into Potential Mechanisms Involved in Aberrant Aggregation in Familial Amyotrophic Lateral Sclerosis.G3 (Bethesda). 2019 Mar 7;9(3):719-728. doi: 10.1534/g3.118.200787. G3 (Bethesda). 2019. PMID: 30622123 Free PMC article.
-
Copper ion / H2O2 oxidation of Cu/Zn-Superoxide dismutase: Implications for enzymatic activity and antioxidant action.Redox Biol. 2019 Sep;26:101262. doi: 10.1016/j.redox.2019.101262. Epub 2019 Jun 28. Redox Biol. 2019. PMID: 31284117 Free PMC article.
-
Dynamical roles of metal ions and the disulfide bond in Cu, Zn superoxide dismutase folding and aggregation.Proc Natl Acad Sci U S A. 2008 Dec 16;105(50):19696-701. doi: 10.1073/pnas.0803266105. Epub 2008 Dec 3. Proc Natl Acad Sci U S A. 2008. PMID: 19052230 Free PMC article.
References
-
- Liochev, S. I. & Fridovich, I. (2003) Free Radical Biol. Med. 34, 1383–1389. - PubMed
-
- Andersen, P. M., Sims, K. B., Xin, W. W., Kiely, R., O'Neill, G., Ravits, J., Pioro, E., Harati, Y., Brower, R. D., Levine, J. S., et al. (2003) Amyotroph. Lateral Scler. Other Motor Neuron Disord. 4, 62–73. - PubMed
-
- Wood, J. D., Beaujeux, T. P. & Shaw, P. J. (2003) Neuropathol. Appl. Neurobiol. 29, 529–545. - PubMed
-
- Dobson, C. M. (2003) Nature 426, 884–890. - PubMed
-
- Kunst, C. B., Mezey, E., Brownstein, M. J. & Patterson, D. (1997) Nat. Genet. 15, 91–94. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

