Components of coated vesicles and nuclear pore complexes share a common molecular architecture
- PMID: 15523559
- PMCID: PMC524472
- DOI: 10.1371/journal.pbio.0020380
Components of coated vesicles and nuclear pore complexes share a common molecular architecture
Erratum in
- PLoS Biol. 2005 Feb;3(2):e80
Abstract
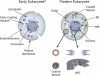
Numerous features distinguish prokaryotes from eukaryotes, chief among which are the distinctive internal membrane systems of eukaryotic cells. These membrane systems form elaborate compartments and vesicular trafficking pathways, and sequester the chromatin within the nuclear envelope. The nuclear pore complex is the portal that specifically mediates macromolecular trafficking across the nuclear envelope. Although it is generally understood that these internal membrane systems evolved from specialized invaginations of the prokaryotic plasma membrane, it is not clear how the nuclear pore complex could have evolved from organisms with no analogous transport system. Here we use computational and biochemical methods to perform a structural analysis of the seven proteins comprising the yNup84/vNup107-160 subcomplex, a core building block of the nuclear pore complex. Our analysis indicates that all seven proteins contain either a beta-propeller fold, an alpha-solenoid fold, or a distinctive arrangement of both, revealing close similarities between the structures comprising the yNup84/vNup107-160 subcomplex and those comprising the major types of vesicle coating complexes that maintain vesicular trafficking pathways. These similarities suggest a common evolutionary origin for nuclear pore complexes and coated vesicles in an early membrane-curving module that led to the formation of the internal membrane systems in modern eukaryotes.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures
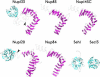


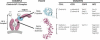
References
-
- Amos LA, van den Ent F, Lowe J. Structural/functional homology between the bacterial and eukaryotic cytoskeletons. Curr Opin Cell Biol. 2004;16:24–31. - PubMed
-
- Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: Structures, functions, and evolution. J Struct Biol. 2001a;134:117–131. - PubMed
-
- Andrade MA, Petosa C, O'Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001b;309:1–18. - PubMed
-
- Bednenko J, Cingolani G, Gerace L. Nucleocytoplasmic transport: Navigating the channel. Traffic. 2003;4:127–135. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

