Formation of membrane-bound ring complexes by prohibitins in mitochondria
- PMID: 15525670
- PMCID: PMC539169
- DOI: 10.1091/mbc.e04-09-0807
Formation of membrane-bound ring complexes by prohibitins in mitochondria
Abstract
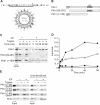
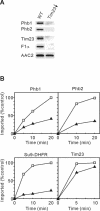
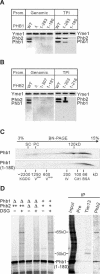
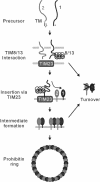
Prohibitins comprise a remarkably conserved protein family in eukaryotic cells with proposed functions in cell cycle progression, senescence, apoptosis, and the regulation of mitochondrial activities. Two prohibitin homologues, Phb1 and Phb2, assemble into a high molecular weight complex of approximately 1.2 MDa in the mitochondrial inner membrane, but a nuclear localization of Phb1 and Phb2 also has been reported. Here, we have analyzed the biogenesis and structure of the prohibitin complex in Saccharomyces cerevisiae. Both Phb1 and Phb2 subunits are targeted to mitochondria by unconventional noncleavable targeting sequences at their amino terminal end. Membrane insertion involves binding of newly imported Phb1 to Tim8/13 complexes in the intermembrane space and is mediated by the TIM23-translocase. Assembly occurs via intermediate-sized complexes of approximately 120 kDa containing both Phb1 and Phb2. Conserved carboxy-terminal coiled-coil regions in both subunits mediate the formation of large assemblies in the inner membrane. Single particle electron microscopy of purified prohibitin complexes identifies diverse ring-shaped structures with outer dimensions of approximately 270 x 200 angstroms. Implications of these findings for proposed cellular activities of prohibitins are discussed.
Figures



References
-
- Arlt, H., Tauer, R., Feldmann, H., Neupert, W., and Langer, T. (1996). The YTA10-12-complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85, 875-885. - PubMed
-
- Artal-Sanz, M., Tsang, W. Y., Willems, E. M., Grivell, L. A., Lemire, B. D., van der Spek, H., Nijtmans, L. G., and Sanz, M. A. (2003). The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans. J. Biol. Chem. 278, 32091-32099. - PubMed
-
- Back, J. W., Sanz, M. A., De Jong, L., De Koning, L. J., Nijtmans, L. G., De Koster, C. G., Grivell, L. A., Van Der Spek, H., and Muijsers, A. O. (2002). A structure for the yeast prohibitin complex: structure prediction and evidence from chemical crosslinking and mass spectrometry. Protein Sci. 11, 2471-2478. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

