Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs
- PMID: 15525708
- PMCID: PMC1370684
- DOI: 10.1261/rna.7135204
Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs
Abstract
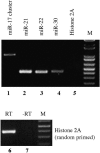
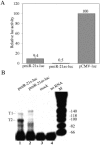
The factors regulating the expression of microRNAs (miRNAs), a ubiquitous family of approximately 22-nt noncoding regulatory RNAs, remain undefined. However, it is known that miRNAs are first transcribed as a largely unstructured precursor, termed a primary miRNA (pri-miRNA), which is sequentially processed in the nucleus, to give the approximately 65-nt pre-miRNA hairpin intermediate, and then in the cytoplasm, to give the mature miRNA. Here we have sought to identify the RNA polymerase responsible for miRNA transcription and to define the structure of a full-length human miRNA. We show that the pri-miRNA precursors for nine human miRNAs are both capped and polyadenylated and report the sequence of the full-length, approximately 3433-nt pri-miR-21 RNA. This pri-miR-21 gene sequence is flanked 5' by a promoter element able to transcribe heterologous mRNAs and 3' by a consensus polyadenylation sequence. Nuclear processing of pri-miRNAs was found to be efficient, thus largely preventing the nuclear export of full-length pri-miRNAs. Nevertheless, an intact miRNA stem-loop precursor located in the 3' UTR of a protein coding gene only moderately inhibited expression of the linked open reading frame, probably because the 3' truncated mRNA could still be exported and expressed. Together, these data show that human pri-miRNAs are not only structurally similar to mRNAs but can, in fact, function both as pri-miRNAs and mRNAs.
Figures



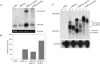

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
