The RNA catabolic enzymes Rex4p, Rnt1p, and Dbr1p show genetic interaction with trans-acting factors involved in processing of ITS1 in Saccharomyces cerevisiae pre-rRNA
- PMID: 15525710
- PMCID: PMC1370683
- DOI: 10.1261/rna.7155904
The RNA catabolic enzymes Rex4p, Rnt1p, and Dbr1p show genetic interaction with trans-acting factors involved in processing of ITS1 in Saccharomyces cerevisiae pre-rRNA
Abstract
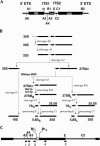
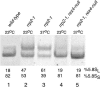
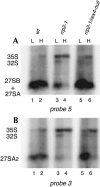
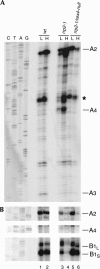
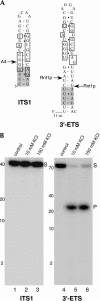
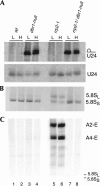
Eukaryotes have two types of ribosomes containing either 5.8SL or 5.8SS rRNA that are produced by alternative pre-rRNA processing. The exact processing pathway for the minor 5.8SL rRNA species is poorly documented. We have previously shown that the trans-acting factor Rrp5p and the RNA exonuclease Rex4p genetically interact to influence the ratio between the two forms of 5.8S rRNA in the yeast Saccharomyces cerevisiae. Here we report a further analysis of ITS1 processing in various yeast mutants that reveals genetic interactions between, on the one hand, Rrp5p and RNase MRP, the endonuclease required for 5.8SS rRNA synthesis, and, on the other, Rex4p, the RNase III homolog Rnt1p, and the debranching enzyme Dbr1p. Yeast cells carrying a temperature-sensitive mutation in RNase MRP (rrp2-1) exhibit a pre-rRNA processing phenotype very similar to that of the previously studied rrp5-33 mutant: ITS2 processing precedes ITS1 processing, 5.8SL rRNA becomes the major species, and ITS1 is processed at the recently reported novel site A4 located midway between sites A2 and A3. As in the rrp5-Delta3 mutant, all of these phenotypical processing features disappear upon inactivation of the REX4 gene. Moreover, inactivation of the DBR1 gene in rrp2-1, or the RNT1 gene in rrp5-Delta3 mutant cells also negates the effects of the original mutation on pre-rRNA processing. These data link a total of three RNA catabolic enzymes, Rex4p, Rnt1p, and Dbr1p, to ITS1 processing and the relative production of 5.8SS and 5.8SL rRNA. A possible model for the indirect involvement of the three enzymes in yeast pre-rRNA processing is discussed.
Figures


Similar articles
-
Deletions in the S1 domain of Rrp5p cause processing at a novel site in ITS1 of yeast pre-rRNA that depends on Rex4p.Nucleic Acids Res. 2002 Oct 1;30(19):4222-31. doi: 10.1093/nar/gkf538. Nucleic Acids Res. 2002. PMID: 12364601 Free PMC article.
-
5'-end formation of yeast 5.8SL rRNA is an endonucleolytic event.Biochem Biophys Res Commun. 2006 Jun 30;345(2):796-802. doi: 10.1016/j.bbrc.2006.04.166. Epub 2006 May 5. Biochem Biophys Res Commun. 2006. PMID: 16701559
-
Ngl2p is a Ccr4p-like RNA nuclease essential for the final step in 3'-end processing of 5.8S rRNA in Saccharomyces cerevisiae.RNA. 2002 Sep;8(9):1095-101. doi: 10.1017/s1355838202021027. RNA. 2002. PMID: 12358428 Free PMC article.
-
Genetic and biochemical analyses of yeast RNase MRP.Mol Biol Rep. 1995-1996;22(2-3):75-9. doi: 10.1007/BF00988709. Mol Biol Rep. 1995. PMID: 8901491 Review.
-
Structure and function of Rnt1p: An alternative to RNAi for targeted RNA degradation.Wiley Interdiscip Rev RNA. 2019 May;10(3):e1521. doi: 10.1002/wrna.1521. Epub 2018 Dec 11. Wiley Interdiscip Rev RNA. 2019. PMID: 30548404 Review.
Cited by
-
The Conserved RNA Exonuclease Rexo5 Is Required for 3' End Maturation of 28S rRNA, 5S rRNA, and snoRNAs.Cell Rep. 2017 Oct 17;21(3):758-772. doi: 10.1016/j.celrep.2017.09.067. Cell Rep. 2017. PMID: 29045842 Free PMC article.
-
Slx9p facilitates efficient ITS1 processing of pre-rRNA in Saccharomyces cerevisiae.RNA. 2006 Nov;12(11):2005-13. doi: 10.1261/rna.159406. Epub 2006 Oct 3. RNA. 2006. PMID: 17018574 Free PMC article.
-
Rrp5p, a trans-acting factor in yeast ribosome biogenesis, is an RNA-binding protein with a pronounced preference for U-rich sequences.RNA. 2006 Feb;12(2):263-71. doi: 10.1261/rna.2257606. RNA. 2006. PMID: 16428605 Free PMC article.
-
Rrp17p is a eukaryotic exonuclease required for 5' end processing of Pre-60S ribosomal RNA.Mol Cell. 2009 Dec 11;36(5):768-81. doi: 10.1016/j.molcel.2009.11.011. Mol Cell. 2009. PMID: 20005841 Free PMC article.
-
Different mechanisms for pseudouridine formation in yeast 5S and 5.8S rRNAs.Mol Cell Biol. 2008 May;28(10):3089-100. doi: 10.1128/MCB.01574-07. Epub 2008 Mar 10. Mol Cell Biol. 2008. PMID: 18332121 Free PMC article.
References
-
- Abou-Elela, S., Igel, H., and Ares Jr., M. 1996. RNAase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85: 115–124. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
