Generation of a catalytic module on a self-folding RNA
- PMID: 15525711
- PMCID: PMC1370678
- DOI: 10.1261/rna.7170304
Generation of a catalytic module on a self-folding RNA
Erratum in
- RNA. 2005 Jan;11(1):114
Abstract
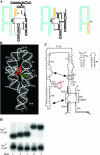
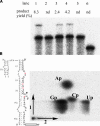
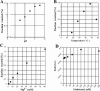
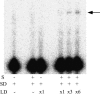
It is theoretically possible to obtain a catalytic site of an artificial ribozyme from a random sequence consisting of a limited numbers of nucleotides. However, this strategy has been inadequately explored. Here, we report an in vitro selection technique that exploits modular construction of a structurally constrained RNA to acquire a catalytic site for RNA ligation from a short random sequence. To practice the selection, a sequence of 30 nucleotides was located close to the putative reaction site in a derivative of a naturally occurring self-folding RNA whose crystal structure is known. RNAs whose activity depended on the starting three-dimensional structure were selected with 3'-5' ligation specificity, indicating that the strategy can be used to acquire a variety of catalytic sites and other functional RNA modules.
Figures


Similar articles
-
A ligase ribozyme obtained from a structured pool.Nucleic Acids Symp Ser (Oxf). 2004;(48):209-10. doi: 10.1093/nass/48.1.209. Nucleic Acids Symp Ser (Oxf). 2004. PMID: 17150552
-
Probing the folding landscape of the Tetrahymena ribozyme: commitment to form the native conformation is late in the folding pathway.J Mol Biol. 2001 May 18;308(5):839-51. doi: 10.1006/jmbi.2001.4751. J Mol Biol. 2001. PMID: 11352576
-
Kinetic and secondary structure analysis of Naegleria andersoni GIR1, a group I ribozyme whose putative biological function is site-specific hydrolysis.Biochemistry. 1997 Dec 23;36(51):16345-54. doi: 10.1021/bi9718595. Biochemistry. 1997. PMID: 9405070
-
Exposing the kinetic traps in RNA folding.Curr Opin Struct Biol. 1999 Jun;9(3):339-45. doi: 10.1016/S0959-440X(99)80045-1. Curr Opin Struct Biol. 1999. PMID: 10361090 Review.
-
RNA tectonics: towards RNA design.Fold Des. 1996;1(4):R78-88. doi: 10.1016/S1359-0278(96)00037-5. Fold Des. 1996. PMID: 9079386 Review.
Cited by
-
Group I introns and inteins: disparate origins but convergent parasitic strategies.J Bacteriol. 2009 Oct;191(20):6193-202. doi: 10.1128/JB.00675-09. Epub 2009 Aug 7. J Bacteriol. 2009. PMID: 19666710 Free PMC article. Review. No abstract available.
-
In vitro RNA random pools are not structurally diverse: a computational analysis.RNA. 2005 Jun;11(6):853-63. doi: 10.1261/rna.7271405. RNA. 2005. PMID: 15923372 Free PMC article.
-
RNA modularity for synthetic biology.F1000Prime Rep. 2013 Nov 1;5:46. doi: 10.12703/P5-46. eCollection 2013. F1000Prime Rep. 2013. PMID: 24273647 Free PMC article. Review.
-
A combinatorial method to isolate short ribozymes from complex ribozyme libraries.Nucleic Acids Res. 2020 Nov 18;48(20):e116. doi: 10.1093/nar/gkaa834. Nucleic Acids Res. 2020. PMID: 33035338 Free PMC article.
-
Emergence of a fast-reacting ribozyme that is capable of undergoing continuous evolution.Proc Natl Acad Sci U S A. 2007 Sep 25;104(39):15288-93. doi: 10.1073/pnas.0707490104. Epub 2007 Sep 18. Proc Natl Acad Sci U S A. 2007. PMID: 17878292 Free PMC article.
References
-
- Bartel, D.P. and Szostak, J.W. 1993. Isolation of new ribozymes from a large pool of random sequences. Science 261: 1411–1418. - PubMed
-
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.l., Kundrot, C.E., Cech, T.R., and Doudna, J.A. 1996. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273: 1678–1685. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
