Generation of a catalytic module on a self-folding RNA
- PMID: 15525711
- PMCID: PMC1370678
- DOI: 10.1261/rna.7170304
Generation of a catalytic module on a self-folding RNA
Erratum in
- RNA. 2005 Jan;11(1):114
Abstract
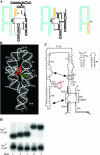
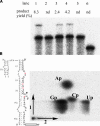
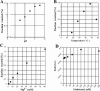
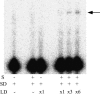
It is theoretically possible to obtain a catalytic site of an artificial ribozyme from a random sequence consisting of a limited numbers of nucleotides. However, this strategy has been inadequately explored. Here, we report an in vitro selection technique that exploits modular construction of a structurally constrained RNA to acquire a catalytic site for RNA ligation from a short random sequence. To practice the selection, a sequence of 30 nucleotides was located close to the putative reaction site in a derivative of a naturally occurring self-folding RNA whose crystal structure is known. RNAs whose activity depended on the starting three-dimensional structure were selected with 3'-5' ligation specificity, indicating that the strategy can be used to acquire a variety of catalytic sites and other functional RNA modules.
Figures


References
-
- Bartel, D.P. and Szostak, J.W. 1993. Isolation of new ribozymes from a large pool of random sequences. Science 261: 1411–1418. - PubMed
-
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.l., Kundrot, C.E., Cech, T.R., and Doudna, J.A. 1996. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273: 1678–1685. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
