Modified nucleotides at the 5' end of human U2 snRNA are required for spliceosomal E-complex formation
- PMID: 15525712
- PMCID: PMC1370681
- DOI: 10.1261/rna.7186504
Modified nucleotides at the 5' end of human U2 snRNA are required for spliceosomal E-complex formation
Abstract
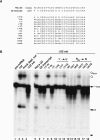
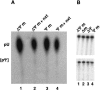
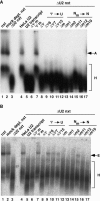
U2 snRNA, a key player in nuclear pre-mRNA splicing, contains a 5'-terminal m3G cap and many internal modifications. The latter were shown in vertebrates to be generally required for U2 function in splicing, but precisely which residues are essential and their role in snRNP and/or spliceosome assembly is presently not clear. Here, we investigated the roles of individual modified nucleotides of HeLa U2 snRNA in pre-mRNA splicing, using a two-step in vitro reconstitution/complementation assay. We show that the three pseudouridines and five 2'O-methyl groups within the first 20 nucleotides of U2 snRNA, but not the m3G cap, are required for efficient pre-mRNA splicing. Individual pseudouridines were not essential, but had cumulative effects on U2 function. In contrast, four of five 2'O-methylations (at positions 1, 2, 12, and 19) were individually required for splicing. The in vitro assembly of 17S U2 snRNPs was not dependent on the presence of modified U2 residues. However, individual internal modifications were required for the formation of the ATP-independent early spliceosomal E complex. Our data strongly suggest that modifications within the first 20 nucleotides of U2 play an important role in facilitating the interaction of U2 with U1 snRNP and/or other factors within the E complex.
Figures
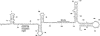

Similar articles
-
The 5' end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes.Mol Cell. 2007 Feb 9;25(3):399-411. doi: 10.1016/j.molcel.2006.12.019. Mol Cell. 2007. PMID: 17289587
-
An RNA-dependent ATPase associated with U2/U6 snRNAs in pre-mRNA splicing.Nature. 1996 Jun 20;381(6584):709-13. doi: 10.1038/381709a0. Nature. 1996. PMID: 8649518
-
Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing.EMBO J. 1998 Oct 1;17(19):5783-95. doi: 10.1093/emboj/17.19.5783. EMBO J. 1998. PMID: 9755178 Free PMC article.
-
Towards understanding the catalytic core structure of the spliceosome.Biochem Soc Trans. 2005 Jun;33(Pt 3):447-9. doi: 10.1042/BST0330447. Biochem Soc Trans. 2005. PMID: 15916538 Review.
-
Splicing of a rare class of introns by the U12-dependent spliceosome.Biol Chem. 2005 Aug;386(8):713-24. doi: 10.1515/BC.2005.084. Biol Chem. 2005. PMID: 16201866 Review.
Cited by
-
Pseudouridine synthase 1 deficient mice, a model for Mitochondrial Myopathy with Sideroblastic Anemia, exhibit muscle morphology and physiology alterations.Sci Rep. 2016 May 20;6:26202. doi: 10.1038/srep26202. Sci Rep. 2016. PMID: 27197761 Free PMC article.
-
Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation.Nucleic Acids Res. 2005 Jun 2;33(10):3133-44. doi: 10.1093/nar/gki630. Print 2005. Nucleic Acids Res. 2005. PMID: 15933208 Free PMC article.
-
Combinatorial regulation of alternative splicing.Biochim Biophys Acta Gene Regul Mech. 2019 Nov-Dec;1862(11-12):194392. doi: 10.1016/j.bbagrm.2019.06.003. Epub 2019 Jul 2. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 31276857 Free PMC article. Review.
-
The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region.Genes Dev. 2011 Mar 1;25(5):440-4. doi: 10.1101/gad.2009411. Genes Dev. 2011. PMID: 21363962 Free PMC article.
-
Depletion of SMN by RNA interference in HeLa cells induces defects in Cajal body formation.Nucleic Acids Res. 2006 May 31;34(10):2925-32. doi: 10.1093/nar/gkl374. Print 2006. Nucleic Acids Res. 2006. PMID: 16738131 Free PMC article.
References
-
- Arnez, J.G. and Steitz, T.A. 1994. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 33: 7560–7567. - PubMed
-
- Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNA by the spliceosome. In The RNA world (eds. R.F. Gestelandet al.), pp 525–560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Charette, M. and Gray, M.W. 2000. Pseudouridine in RNA: What, where, how, and why. IUBMB Life 49: 341–351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
