An enigmatic fourth runt domain gene in the fugu genome: ancestral gene loss versus accelerated evolution
- PMID: 15527507
- PMCID: PMC533870
- DOI: 10.1186/1471-2148-4-43
An enigmatic fourth runt domain gene in the fugu genome: ancestral gene loss versus accelerated evolution
Abstract
Background: The runt domain transcription factors are key regulators of developmental processes in bilaterians, involved both in cell proliferation and differentiation, and their disruption usually leads to disease. Three runt domain genes have been described in each vertebrate genome (the RUNX gene family), but only one in other chordates. Therefore, the common ancestor of vertebrates has been thought to have had a single runt domain gene.
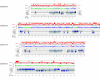
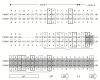
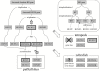
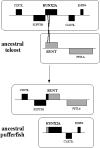
Results: Analysis of the genome draft of the fugu pufferfish (Takifugu rubripes) reveals the existence of a fourth runt domain gene, FrRUNT, in addition to the orthologs of human RUNX1, RUNX2 and RUNX3. The tiny FrRUNT packs six exons and two putative promoters in just 3 kb of genomic sequence. The first exon is located within an intron of FrSUPT3H, the ortholog of human SUPT3H, and the first exon of FrSUPT3H resides within the first intron of FrRUNT. The two gene structures are therefore "interlocked". In the human genome, SUPT3H is instead interlocked with RUNX2. FrRUNT has no detectable ortholog in the genomes of mammals, birds or amphibians. We consider alternative explanations for an apparent contradiction between the phylogenetic data and the comparison of the genomic neighborhoods of human and fugu runt domain genes. We hypothesize that an ancient RUNT locus was lost in the tetrapod lineage, together with FrFSTL6, a member of a novel family of follistatin-like genes.
Conclusions: Our results suggest that the runt domain family may have started expanding in chordates much earlier than previously thought, and exemplify the importance of detailed analysis of whole-genome draft sequence to provide new insights into gene evolution.
Figures




Similar articles
-
Identification of a new pebp2alphaA2 isoform from zebrafish runx2 capable of inducing osteocalcin gene expression in vitro.J Bone Miner Res. 2005 Aug;20(8):1440-53. doi: 10.1359/JBMR.050318. Epub 2005 Mar 21. J Bone Miner Res. 2005. PMID: 16007341
-
cDNA cloning of Runx family genes from the pufferfish (Fugu rubripes).Gene. 2007 Sep 15;399(2):162-73. doi: 10.1016/j.gene.2007.05.014. Epub 2007 Jun 5. Gene. 2007. PMID: 17604919
-
Origin and diversity of the SOX transcription factor gene family: genome-wide analysis in Fugu rubripes.Gene. 2004 Mar 17;328:177-86. doi: 10.1016/j.gene.2003.12.008. Gene. 2004. PMID: 15019997
-
Oncogenic potential of the RUNX gene family: 'overview'.Oncogene. 2004 May 24;23(24):4198-208. doi: 10.1038/sj.onc.1207755. Oncogene. 2004. PMID: 15156173 Review.
-
The Runx genes: lineage-specific oncogenes and tumor suppressors.Oncogene. 2004 May 24;23(24):4308-14. doi: 10.1038/sj.onc.1207130. Oncogene. 2004. PMID: 15156187 Review.
Cited by
-
The vertebrate RCAN gene family: novel insights into evolution, structure and regulation.PLoS One. 2014 Jan 20;9(1):e85539. doi: 10.1371/journal.pone.0085539. eCollection 2014. PLoS One. 2014. PMID: 24465593 Free PMC article.
-
The evolution of Runx genes II. The C-terminal Groucho recruitment motif is present in both eumetazoans and homoscleromorphs but absent in a haplosclerid demosponge.BMC Res Notes. 2009 Apr 17;2:59. doi: 10.1186/1756-0500-2-59. BMC Res Notes. 2009. PMID: 19374764 Free PMC article.
-
Characterization of the Runx gene family in a jawless vertebrate, the Japanese lamprey (Lethenteron japonicum).PLoS One. 2014 Nov 18;9(11):e113445. doi: 10.1371/journal.pone.0113445. eCollection 2014. PLoS One. 2014. PMID: 25405766 Free PMC article.
-
Runx family genes in a cartilaginous fish, the elephant shark (Callorhinchus milii).PLoS One. 2014 Apr 3;9(4):e93816. doi: 10.1371/journal.pone.0093816. eCollection 2014. PLoS One. 2014. PMID: 24699678 Free PMC article.
-
Differential expression of primary pair-rule genes during bidirectional regeneration in Perionyx excavatus.Genes Genomics. 2018 Jul;40(7):747-753. doi: 10.1007/s13258-018-0683-3. Epub 2018 Apr 4. Genes Genomics. 2018. PMID: 29934812
References
-
- Kania MA, Bonner AS, Duffy JB, Gergen JP. The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes Dev. 1990;4:1701–1713. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

