Hydrophobic interactions drive ligand-receptor recognition for activation and inhibition of staphylococcal quorum sensing
- PMID: 15528279
- PMCID: PMC528941
- DOI: 10.1073/pnas.0404039101
Hydrophobic interactions drive ligand-receptor recognition for activation and inhibition of staphylococcal quorum sensing
Abstract
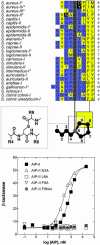
Two-component systems represent the most widely used signaling paradigm in living organisms. Encoding the prototypical two-component system in Gram-positive bacteria, the staphylococcal agr (accessory gene regulator) operon uses a polytopic receptor, AgrC, activated by an autoinducing peptide (AIP), to coordinate quorum sensing with the global synthesis of virulence factors. The agr locus has undergone evolutionary divergence, resulting in the formation of several distinct inter- and intraspecies specificity groups, such that most cross-group AIP-receptor interactions are mutually inhibitory. We have exploited this natural diversity by constructing and analyzing AgrC chimeras generated by exchange of intradomain segments between receptors of different agr groups. Functional chimeras fell into three general classes: receptors with broadened specificity, receptors with tightened specificity, and receptors that lack activation specificity. Testing of these chimeric receptors against a battery of AIP analogs localized the primary ligand recognition site to the receptor distal subdomain and revealed that the AIPs bind primarily to a putative hydrophobic pocket in the receptor. This binding is mediated by a highly conserved hydrophobic patch on the AIPs and is an absolute requirement for interactions in self-activation and cross-inhibition of the receptors. It is suggested that this recognition scheme provides the fundamental basis for agr activation and interference.
Figures



References
-
- Novick, R. P. & Muir, T. W. (1999) Curr. Opin. Microbiol. 2, 40-45. - PubMed
-
- Kleerebezem, M., Quadri, L. E., Kuipers, O. P. & de Vos, W. M. (1997) Mol. Microbiol. 24, 895-904. - PubMed
-
- Nakayama, J., Cao, Y., Horii, T., Sakuda, S., Akkermans, A. D., de Vos, W. M. & Nagasawa, H. (2001) Mol. Microbiol. 41, 145-154. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

