Aflatoxin biosynthesis cluster gene cypA is required for G aflatoxin formation
- PMID: 15528514
- PMCID: PMC525170
- DOI: 10.1128/AEM.70.11.6518-6524.2004
Aflatoxin biosynthesis cluster gene cypA is required for G aflatoxin formation
Abstract
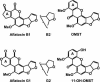
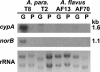
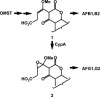
Aspergillus flavus isolates produce only aflatoxins B1 and B2, while Aspergillus parasiticus and Aspergillus nomius produce aflatoxins B1, B2, G1, and G2. Sequence comparison of the aflatoxin biosynthesis pathway gene cluster upstream from the polyketide synthase gene, pksA, revealed that A. flavus isolates are missing portions of genes (cypA and norB) predicted to encode, respectively, a cytochrome P450 monooxygenase and an aryl alcohol dehydrogenase. Insertional disruption of cypA in A. parasiticus yielded transformants that lack the ability to produce G aflatoxins but not B aflatoxins. The enzyme encoded by cypA has highest amino acid identity to Gibberella zeae Tri4 (38%), a P450 monooxygenase previously shown to be involved in trichodiene epoxidation. The substrate for CypA may be an intermediate formed by oxidative cleavage of the A ring of O-methylsterigmatocystin by OrdA, the P450 monooxygenase required for formation of aflatoxins B1 and B2.
Figures



Similar articles
-
Characterization of the critical amino acids of an Aspergillus parasiticus cytochrome P-450 monooxygenase encoded by ordA that is involved in the biosynthesis of aflatoxins B1, G1, B2, and G2.Appl Environ Microbiol. 1998 Dec;64(12):4834-41. doi: 10.1128/AEM.64.12.4834-4841.1998. Appl Environ Microbiol. 1998. PMID: 9835571 Free PMC article.
-
Genes encoding cytochrome P450 and monooxygenase enzymes define one end of the aflatoxin pathway gene cluster in Aspergillus parasiticus.Appl Microbiol Biotechnol. 2000 May;53(5):583-90. doi: 10.1007/s002530051660. Appl Microbiol Biotechnol. 2000. PMID: 10855719
-
aflT, a MFS transporter-encoding gene located in the aflatoxin gene cluster, does not have a significant role in aflatoxin secretion.Fungal Genet Biol. 2004 Oct;41(10):911-20. doi: 10.1016/j.fgb.2004.06.007. Fungal Genet Biol. 2004. PMID: 15341913
-
Understanding nonaflatoxigenicity of Aspergillus sojae: a windfall of aflatoxin biosynthesis research.Appl Microbiol Biotechnol. 2007 Oct;76(5):977-84. doi: 10.1007/s00253-007-1116-4. Epub 2007 Jul 31. Appl Microbiol Biotechnol. 2007. PMID: 17665189 Review.
-
Molecular genetic analysis and regulation of aflatoxin biosynthesis.Appl Microbiol Biotechnol. 2003 Apr;61(2):83-93. doi: 10.1007/s00253-002-1199-x. Epub 2003 Jan 28. Appl Microbiol Biotechnol. 2003. PMID: 12655449 Review.
Cited by
-
Identification of an intermediate for 1,8-dihydroxynaphthalene-melanin synthesis in a race-2 isolate of Fulvia fulva (syn. Cladosporium fulvum).Heliyon. 2018 Dec 17;4(12):e01036. doi: 10.1016/j.heliyon.2018.e01036. eCollection 2018 Dec. Heliyon. 2018. PMID: 30582052 Free PMC article.
-
Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species.PLoS Biol. 2017 Nov 17;15(11):e2003583. doi: 10.1371/journal.pbio.2003583. eCollection 2017 Nov. PLoS Biol. 2017. PMID: 29149178 Free PMC article.
-
First record of Aspergillus nomiae as a broad-spectrum entomopathogenic fungus that provides resistance against phytopathogens and insect pests by colonization of plants.Front Microbiol. 2024 Jan 8;14:1284276. doi: 10.3389/fmicb.2023.1284276. eCollection 2023. Front Microbiol. 2024. PMID: 38260878 Free PMC article.
-
Degeneration of aflatoxin gene clusters in Aspergillus flavus from Africa and North America.AMB Express. 2016 Dec;6(1):62. doi: 10.1186/s13568-016-0228-6. Epub 2016 Aug 31. AMB Express. 2016. PMID: 27576895 Free PMC article.
-
Gene duplication, modularity and adaptation in the evolution of the aflatoxin gene cluster.BMC Evol Biol. 2007 Jul 9;7:111. doi: 10.1186/1471-2148-7-111. BMC Evol Biol. 2007. PMID: 17620135 Free PMC article.
References
-
- Bayman, P., and P. J. Cotty. 1991. Improved media for selecting nitrate-nonutilizing mutants in Aspergillus flavus. Mycologia 83:311-316.
-
- Bhatnagar, D., T. E. Cleveland, and D. G. Kingston. 1991. Enzymological evidence for separate pathways for aflatoxin B1 and B2 biosynthesis. Biochemistry 30:4343-4350. - PubMed
-
- Bhatnagar, D., K. C. Ehrlich, and T. E. Cleveland. 2003. Molecular genetic analysis and regulation of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 61:83-93. - PubMed
-
- Bhatnagar, D., K. C. Ehrlich, and T. E. Cleveland. 1992. Oxidation-reduction reactions in biosynthesis of secondary metabolites, p. 255-285. In D. Bhatnagar, E. B. Lillehoj, and D. K. Arora (ed.), Mycotoxins in ecological systems, vol. 10. Marcel Dekker, New York, N.Y.
-
- Cary, J. W., J. M. Dyer, K. C. Ehrlich, M. S. Wright, S.-H. Liang, and J. E. Linz. 2002. Molecular and functional characterization of a second copy of the aflatoxin regulatory gene, aflR-2, from Aspergillus parasiticus. Biochim. Biophys. Acta 1576:316-323. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

