Diversity and antagonistic potential of bacteria associated with bryophytes from nutrient-poor habitats of the Baltic Sea Coast
- PMID: 15528520
- PMCID: PMC525113
- DOI: 10.1128/AEM.70.11.6569-6579.2004
Diversity and antagonistic potential of bacteria associated with bryophytes from nutrient-poor habitats of the Baltic Sea Coast
Abstract
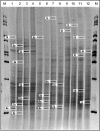
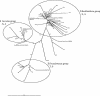
Very little is known about the interaction of bryophytes with bacteria. Therefore, we analyzed bacteria associated with three bryophyte species, Tortula ruralis, Aulacomnium palustre, and Sphagnum rubellum, which represent typical moss species of three nutrient-poor plant communities at the southern Baltic Sea coast in Germany. By use of two cultivation-independent techniques, denaturing gradient gel electrophoresis and single-strand conformation polymorphism analysis of the 16S ribosomal DNA, a high degree of moss specificity was found for associated bacterial communities. This specificity could be further evidenced by a cultivation-dependent approach for the following parameters: (i) plate counts of bacteria on R2A medium, (ii) proportion of antagonistic isolates, (iii) antagonistic activity as well as spectrum against pathogens, and (iv) diversity and richness of antagonistic isolates. The proportion of isolates with antagonistic activity against the pathogenic model fungus Verticillium dahliae was highest for S. rubellum (31%), followed by A. palustre (17%) and T. ruralis (5%). A high percentage (99%) of moss-associated antagonistic bacteria produced antifungal compounds. The high recovery of antagonistic isolates strongly suggests that bryophytes represent an ecological niche which harbors a diverse and hitherto largely uncharacterized microbial population with yet unknown and untapped potential biotechnological applications, e.g., for biological control of plant pathogens.
Figures



Similar articles
-
Potato-associated bacteria and their antagonistic potential towards plant-pathogenic fungi and the plant-parasitic nematode Meloidogyne incognita (Kofoid & White) Chitwood.Can J Microbiol. 2002 Sep;48(9):772-86. doi: 10.1139/w02-071. Can J Microbiol. 2002. PMID: 12455609
-
The bryophyte genus Sphagnum is a reservoir for powerful and extraordinary antagonists and potentially facultative human pathogens.FEMS Microbiol Ecol. 2007 Jul;61(1):38-53. doi: 10.1111/j.1574-6941.2007.00323.x. Epub 2007 Apr 30. FEMS Microbiol Ecol. 2007. PMID: 17484734
-
Impact of plant species and site on rhizosphere-associated fungi antagonistic to Verticillium dahliae kleb.Appl Environ Microbiol. 2005 Aug;71(8):4203-13. doi: 10.1128/AEM.71.8.4203-4213.2005. Appl Environ Microbiol. 2005. PMID: 16085804 Free PMC article.
-
Burkholderia bryophila sp. nov. and Burkholderia megapolitana sp. nov., moss-associated species with antifungal and plant-growth-promoting properties.Int J Syst Evol Microbiol. 2007 Oct;57(Pt 10):2228-2235. doi: 10.1099/ijs.0.65142-0. Int J Syst Evol Microbiol. 2007. PMID: 17911288
-
Endophytic bacterial communities of field-grown potato plants and their plant-growth-promoting and antagonistic abilities.Can J Microbiol. 2004 Apr;50(4):239-49. doi: 10.1139/w03-118. Can J Microbiol. 2004. PMID: 15213748
Cited by
-
Long-Term Recovery of Microbial Communities in the Boreal Bryosphere Following Fire Disturbance.Microb Ecol. 2017 Jan;73(1):75-90. doi: 10.1007/s00248-016-0832-7. Epub 2016 Aug 18. Microb Ecol. 2017. PMID: 27538873
-
Specific detection and real-time PCR quantification of potentially mycophagous bacteria belonging to the genus Collimonas in different soil ecosystems.Appl Environ Microbiol. 2007 Jul;73(13):4191-7. doi: 10.1128/AEM.00387-07. Epub 2007 May 4. Appl Environ Microbiol. 2007. PMID: 17483278 Free PMC article.
-
Mosses as extraordinary reservoir of microbial diversity: a comparative analysing of co-occurring 'plant-moss twins' in natural alpine ecosystem.Environ Microbiome. 2025 Jun 4;20(1):61. doi: 10.1186/s40793-025-00728-z. Environ Microbiome. 2025. PMID: 40468449 Free PMC article.
-
Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants.Front Microbiol. 2014 Feb 19;5:64. doi: 10.3389/fmicb.2014.00064. eCollection 2014. Front Microbiol. 2014. PMID: 24600444 Free PMC article.
-
Cultivating uncultured bacteria from northern wetlands: knowledge gained and remaining gaps.Front Microbiol. 2011 Sep 16;2:184. doi: 10.3389/fmicb.2011.00184. eCollection 2011. Front Microbiol. 2011. PMID: 21954394 Free PMC article.
References
-
- Ando, H., and A. Mastuo. 1984. Applied bryology. Adv. Bryol. 2:133-224.
-
- Bassam, B. J., G. Ceatano-Anolles, and P. M. Gresshoff. 2001. Fast and sensitive silver staining of DNA in polyacylamide gels. Anal. Biochem. 196:80-83. - PubMed
-
- Berg, G. 1996. Rhizobacteria of oil seed rape antagonistic to Verticillium dahliae. J. Plant Dis. Prot. 103:20-30.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

