Life history implications of rRNA gene copy number in Escherichia coli
- PMID: 15528533
- PMCID: PMC525164
- DOI: 10.1128/AEM.70.11.6670-6677.2004
Life history implications of rRNA gene copy number in Escherichia coli
Abstract
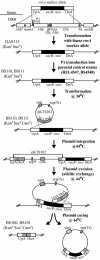
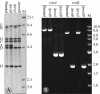
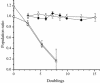
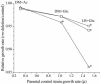
The role of the rRNA gene copy number as a central component of bacterial life histories was studied by using strains of Escherichia coli in which one or two of the seven rRNA operons (rrnA and/or rrnB) were deleted. The relative fitness of these strains was determined in competition experiments in both batch and chemostat cultures. In batch cultures, the decrease in relative fitness corresponded to the number of rRNA operons deleted, which could be accounted for completely by increased lag times and decreased growth rates. The magnitude of the deleterious effect varied with the environment in which fitness was measured: the negative consequences of rRNA operon deletions increased under culture conditions permitting more-rapid growth. The rRNA operon deletion strains were not more effective competitors under the regimen of constant, limited resources provided in chemostat cultures. Enhanced fitness in chemostat cultures would have suggested a simple tradeoff in which deletion strains grew faster (due to more efficient resource utilization) under resource limitation. The contributions of growth rate, lag time, Ks, and death rate to the fitness of each strain were verified through mathematical simulation of competition experiments. These data support the hypothesis that multiple rRNA operons are a component of bacterial life history and that they confer a selective advantage permitting microbes to respond quickly and grow rapidly in environments characterized by fluctuations in resource availability.
Figures
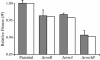
References
-
- Bachellier, S., E. Gilson, M. Hofnung, and C. W. Hill. 1996. Repeated sequences, p. 2012-2040. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. American Society for Microbiology, Washington, D.C.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

