Use of cell culture-PCR assay based on combination of A549 and BGMK cell lines and molecular identification as a tool to monitor infectious adenoviruses and enteroviruses in river water
- PMID: 15528536
- PMCID: PMC525112
- DOI: 10.1128/AEM.70.11.6695-6705.2004
Use of cell culture-PCR assay based on combination of A549 and BGMK cell lines and molecular identification as a tool to monitor infectious adenoviruses and enteroviruses in river water
Abstract
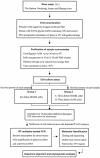
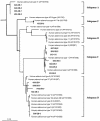
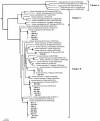
Viral contamination in environmental samples can be underestimated because a single cell line might reproduce only some enteric viruses and some enteric viruses do not exhibit apparent cytopathic effects in cell culture. To overcome this problem, we evaluated a cell culture-PCR assay based on a combination of A549 and Buffalo green monkey kidney (BGMK) cell lines as a tool to monitor infectious adenoviruses and enteroviruses in river water. Water samples were collected 10 times at each of four rivers located in Gyeonggi Province, South Korea, and then cultured on group 1 cells (BGMK cells alone) and group 2 cells (BGMK and A549 cells). Reverse transcription and multiplex PCR were performed, followed by phylogenetic analysis of the amplicons. Thirty (75.0%) of the 40 samples were positive for viruses based on cell culture, and the frequency of positive samples grown on group 2 cells (65.0%) was higher than the frequency of positive samples grown on group 1 cells (50.0%). The number of samples positive for adenoviruses was higher with A549 cells (13 samples) than with BGMK cells (one sample); the numbers of samples positive for enteroviruses were similar with both types of cells. By using phylogenetic analysis, adenoviral amplicons were grouped into subgenera A, C, D, and F, and enteroviral amplicons were grouped into coxsackieviruses B3 and B4 and echoviruses 6, 7, and 30, indicating that A549 and BGMK cells were suitable for recovering a wide range of adenoviral and enteroviral types. The cell culture-PCR assay with a combination of A549 and BGMK cells and molecular identification could be a useful tool for monitoring infectious adenoviruses and enteroviruses in aquatic environments.
Figures
References
-
- Allard, A., B. Albinsson, and G. Wadell. 1992. Detection of adenoviruses in stools from healthy persons and patients with diarrhea by two-step polymerase chain reaction. J. Med. Virol. 37:149-157. - PubMed
-
- American Public Health Association. 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Washington, D.C.
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

