Engineering of chimeric class II polyhydroxyalkanoate synthases
- PMID: 15528546
- PMCID: PMC525123
- DOI: 10.1128/AEM.70.11.6789-6799.2004
Engineering of chimeric class II polyhydroxyalkanoate synthases
Abstract
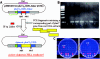
PHA synthase is a key enzyme involved in the biosynthesis of polyhydroxyalkanoates (PHAs). Using a combinatorial genetic strategy to create unique chimeric class II PHA synthases, we have obtained a number of novel chimeras which display improved catalytic properties. To engineer the chimeric PHA synthases, we constructed a synthetic phaC gene from Pseudomonas oleovorans (phaC1Po) that was devoid of an internal 540-bp fragment. Randomly amplified PCR products (created with primers based on conserved phaC sequences flanking the deleted internal fragment) were generated using genomic DNA isolated from soil and were substituted for the 540-bp internal region. The chimeric genes were expressed in a PHA-negative strain of Ralstonia eutropha, PHB(-)4 (DSM 541). Out of 1,478 recombinant clones screened for PHA production, we obtained five different chimeric phaC1Po genes that produced more PHA than the native phaC1Po. Chimeras S1-71, S4-8, S5-58, S3-69, and S3-44 exhibited 1.3-, 1.4-, 2.0-, 2.1-, and 3.0-fold-increased levels of in vivo activity, respectively. All of the mutants mediated the synthesis of PHAs with a slightly increased molar fraction of 3-hydroxyoctanoate; however, the weight-average molecular weights (Mw) of the PHAs in all cases remained almost the same. Based upon DNA sequence analyses, the various phaC fragments appear to have originated from Pseudomonas fluorescens and Pseudomonas aureofaciens. The amino acid sequence analyses showed that the chimeric proteins had 17 to 20 amino acid differences from the wild-type phaC1Po, and these differences were clustered in the same positions in the five chimeric clones. A threading model of PhaC1Po, developed based on homology of the enzyme to the Burkholderia glumae lipase, suggested that the amino acid substitutions found in the active chimeras were located mostly on the protein model surface. Thus, our combinatorial genetic engineering strategy proved to be broadly useful for improving the catalytic activities of PHA synthase enzymes.
Figures



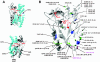
References
-
- Clemente, T., D. Shah, M. Tran, D. Stark, S. Padgette, D. Dennis, K. Brückener, A. Steinbüchel, and T. A. Mitsky. 2000. Sequence of PHA synthase gene from two strains of Rhodospirillum rubrum and in vivo substrate specificity of four PHA synthases across two heterologous expression systems. Appl. Microbiol. Biotechnol. 53:420-429. - PubMed
-
- Green, P. R., J. Kemper, L. Schechtman, L. Guo, M. Satkowski, S. Fiedler, A. Steinbüchel, and B. H. A. Rehm. 2002. Formation of short chain length/medium chain length polyhydroxyalkanoate copolymers by fatty acid β-oxidation inhibited Ralstonia eutropha. Biomacromolecules 3:208-213. - PubMed
-
- Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams. 1993. Bergey's manual of determinative bacteriology, 9th ed. Williams & Wilkins, Baltimore, Md.
-
- Jia, Y., T. J. Kappock, T. Frick, A. J. Sinskey, and J. Stubbe. 2000. Lipases provide a new mechanistic model for polyhydroxybutyrate (PHB) synthases: characterization of the functional residues in Chromatium vinosum PHB synthase. Biochemistry 39:3927-3936. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

