New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria
- PMID: 15528558
- PMCID: PMC525206
- DOI: 10.1128/AEM.70.11.6887-6891.2004
New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria
Abstract
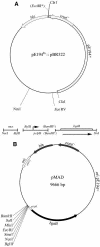
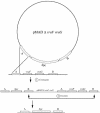
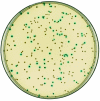
A shuttle vector designated pMAD was constructed for quickly generating gene inactivation mutants in naturally nontransformable gram-positive bacteria. This vector allows, on X-Gal (5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside) plates, a quick colorimetric blue-white discrimination of bacteria which have lost the plasmid, greatly facilitating clone identification during mutagenesis. The plasmid was used in Staphylococcus aureus, Listeria monocytogenes, and Bacillus cereus to efficiently construct mutants with or without an associated antibiotic resistance gene.
Figures
References
-
- Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. - PubMed
-
- Chambers, S. P., S. E. Prior, D. A. Barstow, and N. P. Minton. 1988. The pMTL nic− cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene 68:139-149. - PubMed
-
- Chastanet, A., J. Fert, and T. Msadek. 2003. Comparative genomics reveal novel heat-shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol. Microbiol. 47:1061-1073. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

